Abstract
Identification of genomic signatures that help reveal mechanisms underlying desirable traits in domesticated pigs is of significant biological, agricultural and medical importance. To identify the genomic footprints left by selection during domestication of the Enshi black pig, a typical native and meat-lard breed in China, we generated about 72-fold coverage of the pig genome using pools of genomic DNA representing three different populations of Enshi black pigs from three different locations. Combining this data with the available whole genomes of 13 Chinese wild boars, we identified 417 protein-coding genes embedded in the selected regions of Enshi black pigs. These genes are mainly involved in developmental and metabolic processes, response to stimulus, and other biological processes. Signatures of selection were detected in genes involved in body size and immunity (RPS10 and VASN), lipid metabolism (GSK3), male fertility (INSL6) and developmental processes (TBX19). These findings provide a window into the potential genetic mechanism underlying development of desirable phenotypes in Enshi black pigs during domestication and subsequent artificial selection. Thus, our results illustrate how domestication has shaped patterns of genetic variation in Enshi black pigs and provide valuable genetic resources that enable effective use of pigs in agricultural production.
From early domestication to modern breeding practices, artificial selection for agriculturally important traits has shaped the genomes of domestic pigs. Guided by the reference genome of the domestic Duroc pig, considerable attempts have been made to unveil the genetic bases of phenotypes by using whole-genome wide SNP chip and resequencing approaches1,2,3,4. Wilkinson et al. found genomic regions associated with coat colour and ear morphology using Porcine60 K SNP chip4. Li et al. revealed strong signatures of selection in Berkshire affecting disease resistance, pork yield, fertility, tameness and body length1. Rubin et al. determined three selected genes that associate with body length in European commercial breeds2. Wang et al. revealed evidence of artificial selection of coat colour and reproductive traits in Chinese Tongchen pigs3.
Enshi black pigs, which comprise a typical native black breed in China, are best known for their fat storage ability and cold-wet tolerance. They are mainly raised in mountainous regions with an average altitude of 800 m above sea level in southwest China5 (Enshi Tujia minority and Miao minority autonomous prefecture) (Fig. 1). Enshi black pigs have undergone fewer systematic genetic improvement programs compared to other breeds since the 17th century and are characterized by their average-sized head, concave and wrinkled face, well-developed limbs, concave back, tilted haunch and big belly6. Although they have adapted to the harsh local environment, Enshi black pigs are markedly different from wild boars, especially in terms of fat storage ability. Historically, Enshi black pigs have been intensively used in dry-cured ham production, leading to preferential selection based on their meat and fat quality traits, such as lean percentage, fat content, and eating quality5,6 (juiciness, flavor, tenderness, pink hue and heavy marbling) (Fig. 2).
Figure 1. Geographical distribution of the Enshi black pigs used in our study.
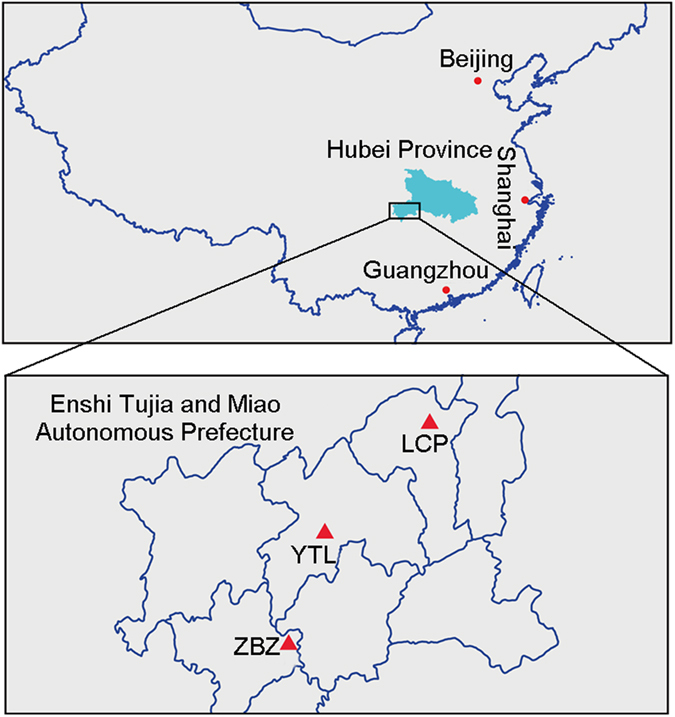
The map was created by using the “maptools” package (v0.839) of R software (v3.1.3, www.r-project.org).
Figure 2. Image of Enshi black pig and Chinese wild boar.
To identify signatures of selection resulting from domestication, we performed whole-genome pooled resequencing of three representative populations of Enshi black pigs (~202.21 Gb in total; ~24.07× coverage per population). Together with 96 publicly available genomes of pig breeds found in Asia (13 Chinese wild boars, 10 Korean wild boars and 73 Chinese domestic pigs) and using a total of ~4.00 trillion bases of resequencing data7,8,9,10,11 (Supplementary Table S1), we conducted a comprehensive analysis of genetic diversity and sought to identify genomic regions under selection in the Enshi black pig.
Results and Discussion
Sequencing, SNP calling and annotation
Pooled paired-end sequencing of the three populations of Enshi black pigs generated 70.64, 67.86 and 63.71 Gb data for the genomes of the Lvcongpo (LCP), Yetinglu (YTL) and Zhongbaozhen (ZBZ) populations, respectively. To accurately detect genomic footprints left by selection, we downloaded the publicly available genome data of 13 Chinese wild boars (Supplementary Table S1). To reduce possible bias resulting from differences in sequencing coverage between the Enahi black pigs (~24.07× coverage per population) and the Chinese wild boars (~13.19× coverage per individual) and to accurately detect genomic footprints left by selection, we randomly selected ~4.80 Gb of high-quality sequencing data from each individual of 13 Chinese wild boars7,9,10, which simulated pooled sequencing of a Chinese wild boar population, yielding a total of 61.82 Gb of sequencing data.
A total of 248.91 Gb high-quality sequencing data from three Enshi black pig populations and a Chinese wild boar population were aligned against the reference pig genome (Sscrofa10.2) using BWA (v0.7.8)12. For each population, ~94.29% of the high-quality reads were mapped to the reference pig genome, of which ~81.82% were uniquely mapped, with comparable genome coverage (~13.32× to 15.70×) between the three populations of Enshi black pig and one population of Chinese wild boar (Supplementary Table S2).
We identified ~14.09 M SNPs in four populations that were concurrently cataloged by two currently dominant algorithms (i.e. SAMtools13 and GATK14), which accounted for 88.12% and 86.76% of SNPs that were identified by SAMtools and GATK, respectively, and were used for subsequent analyses (Supplementary Figure S1, Supplementary Table S3). In total, 0.28–0.30 M (2.01–2.06%) SNPs were considered novel based on their absence in the pig dbSNP (Build 143) database (Supplementary Table S3).
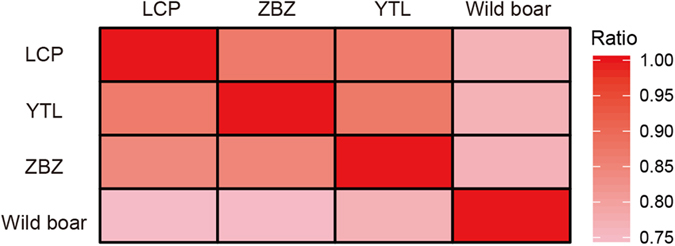
Compared with the 60.92% SNPs (~10.70 M of 17.57 M) that were shared between the Enshi black pigs and Chinese wild boar, more than three-quarter of SNPs (~12.06 M of 15.98 M, or 75.56%) were shared among three Enshi black pig populations (Supplementary Figure S2), which indicated substantial genomic similarity between Enshi black pigs from the three major distributed locations (Fig. 3). Furthermore, the neighbor-joining tree of the four populations revealed two clusters (three Enshi black pig populations and a Chinese wild boar population) (Supplementary Figure S3a). This finding is consistent with the low diversity among the three Enshi black pig populations, as revealed by the phylogenetic analysis of complete mitochondrial DNA sequences of Eurasian pigs (Supplementary Figure S4).
Figure 3. The ratio of shared SNPs between different populations.
We identified 113,864 non-redundant coding SNPs from four populations, of which 91,353, 91,403, 91,639 and 79,155 are from the LCP, YTL, ZBZ and wild boar populations, respectively (Supplementary Figure S5). We detected 76,168 shared coding SNPs in three Enshi black pig populations, and 25,816 nonsynonymous nucleotide substitutions (25,595 missense, 183 stop gain and 38 stop loss) in 8,301 genes (Table 1). The top 1,053 genes containing the highest number (n ≥ 7) of nonsynonymous SNPs were mainly over-represented in the highly variable sensory perception categories (Supplementary Table S4), which reflects the strong reliance of pigs on their sense of smell while scavenging for food and other odor-driven behavior.
Table 1. Summary and annotation of SNPs in Enshi black pigs and Chines wild boar.
| Number of SNPs | |||||
|---|---|---|---|---|---|
| Category | LCP | YTL | ZBZ | ES | Wild boar |
| 3’UTR | 54,883 | 55,078 | 55,961 | 44,505 | 52,282 |
| Intergenic | 9,925,783 | 9,934,711 | 10,260,959 | 7,823,054 | 10,327,917 |
| Splicing | 402 | 421 | 430 | 351 | 392 |
| Upstream | 115,480 | 115,907 | 117,787 | 93,785 | 108,071 |
| Exonic | |||||
| Stop gain | 231 | 231 | 244 | 183 | 221 |
| Nonsynonymous | 30,849 | 30,819 | 31,005 | 25,595 | 26,568 |
| Unknown | 1,325 | 1,312 | 1,352 | 1,133 | 1,149 |
| Synonymous | 60,015 | 60,094 | 60,111 | 50,161 | 52,160 |
| Stop loss | 43 | 42 | 44 | 38 | 41 |
| Downstream | 113,563 | 113,845 | 116,124 | 90,862 | 110,907 |
| 5’UTR | 11,869 | 12,041 | 11,920 | 10,034 | 8,700 |
| Intronic | 3,554,718 | 3,553,004 | 3,642,593 | 2,823,962 | 3,569,327 |
The package ANNOVAR was used to identify whether a SNP causes protein coding changes and the amino acids that are affected. LCP, Lvcongpo population; YTL, Yetinglu population; ZBZ, Zhongbaozhen population; Wild boar, Chinese wild boar population; ES, shared in three Enshi pig populations.
Selective sweep analysis
We analyzed the pooled sequences for selective sweeps in Enshi black pigs by searching for genomic regions with excess homozygosity and/or increased genetic distance to Chinese wild boar (FST). We identified 1,051–1,078 regions (195.15 M to 198.16 M) in the Enshi black pig genome with extremely low levels of heterozygosity (Z(HP) < −2) and 1,043–1,067 regions (208.94 M to 210.96 M) with strongly elevated FST values (Z(FST) > 2). In total, 463–487 unique candidate domestication regions (CDRs) (from 81.77 M to 87.52 M) containing 858–1,046 genes were identified in three populations by focusing our analysis of putatively selected regions on those that fall at least two standard deviations away from the mean (Z(HP) < −2 and Z(FST) > 2, Supplementary Figure S6–8). The large overlap between the HP and FST regions indicated that the two statistical methods detected the same events. SNPs from these CDRs formed two distinct clusters (i.e. three Enshi black pig populations and a Chinese wild boar population) (Supplementary Figure S3b).
We detected 417 candidate selected genes (CSGs) in 185 CDRs (30.91 M, Supplementary Data S1), which were shared among the three populations. Most of the selected genes presented a lower degree of haplotype sharing between the Enshi black pigs and the Chinese wild boar breeds, and highly similarity of haplotypes among three Enshi black pig populations (Fig. 4). These CSGs were mainly overrepresented in developmental processes (hemopoiesis, P = 1.39 × 10−11; cell differentiation, P = 2.27 × 10−9; system development, P = 3.36 × 10−2), metabolic processes (regulation of phosphate metabolic process, P = 7.68 × 10−10; protein phosphorylation, P = 3.47 × 10−7; biosynthetic process, P = 4.04 × 10−4; phosphate-containing compound metabolic process, P = 1.44 × 10−4; and cellular protein modification, P = 8.34 × 10−3), and response to stimulus (natural killer cell activation, P = 4.76 × 10−11; response to stress, P = 1.63 × 10−4) (Table 2). These rapidly evolved genes in Enshi black pigs may be responsible for the dramatic phenotypic changes that are of economic value, such as growth rate, fat storage ability and disease resistance. Six CSGs were found to be related to lipid transport and metabolism (CYP2F1, PISD, TKTL2, SEC14L5, PRSS33, and ACSF2), which may have resulted from selection driven by a demand for energy-rich food during the development of fatty Enshi black pigs in China. Consistent with previous reports1,15 on domesticated pigs, the rapid evolution of 29 genes involved in immune-related processes (Table 2) may have contributed to the human-desirable selection of disease resistance. In addition, a concise FI network was constructed based on the 417 CSGs, with UBC, JAK3, RAC1, RELA and MAPK1 as the central node genes (Supplementary Figure S9). The central node genes were those involved primarily in cellular and immune-related processes.
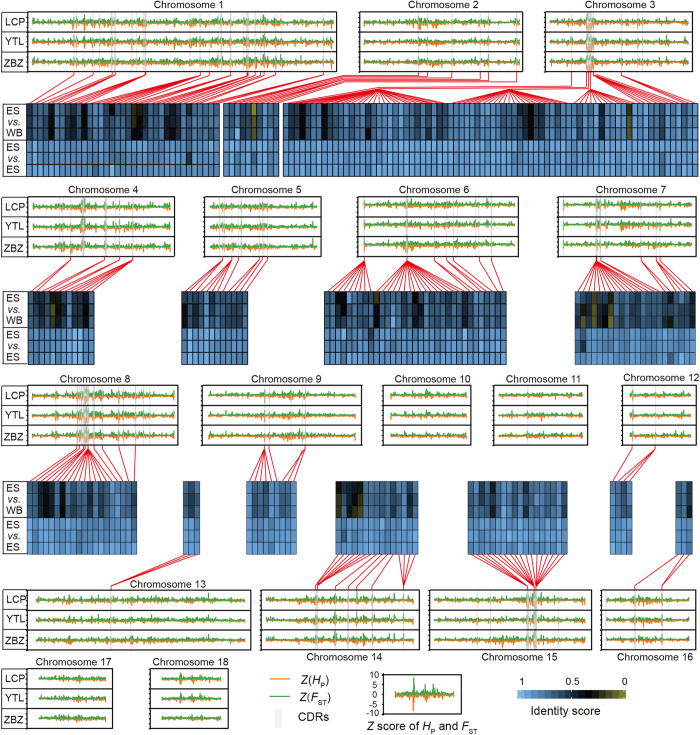
Figure 4. The candidate domestication regions and genes distribution along pig autosomes 1–18.
The positive end of the Z(FST) and the negative end of the Z(HP) distribution plotted for SNPs within each 50-kb window across the genome, the cut-off (|Z| > 2) used for extracting outliers, and the candidate domestication regions shared among three populations were marked with gray shadow. The degree of haplotype sharing for the candidate selected genes (CSGs) were achieved in pairwise comparisons. Boxes to the left indicate the comparison presented on that row (ES, Enshi black pig; WB, Chinese wild boar). Heatmap colors indicate identity scores (IS). LCP, Lvcongpo population; YTL, Yetinglu population; ZBZ, Zhongbaozhen population.
Table 2. Enriched gene ontology terms among CDR genes.
| Term ID | Term description | Gene count | P value |
|---|---|---|---|
| GO-BP:0030101‡ | Natural killer cell activation | 14 | 4.76 × 10−11 |
| GO-BP:0008283 | Cell proliferation | 14 | 8.05 × 10−11 |
| GO-BP:0030097† | Hemopoiesis | 15 | 1.39 × 10−11 |
| GO-BP:0030154† | Cell differentiation | 15 | 2.27 × 10−09 |
| GO-BP:0019220* | Regulation of phosphate metabolic process | 16 | 7.68 × 10−10 |
| GO-BP:0006468* | Protein phosphorylation | 23 | 3.47 × 10−07 |
| GO-BP:0006950‡ | Response to stress | 20 | 1.63 × 10−04 |
| GO-BP:0009058* | Biosynthetic process | 21 | 4.04 × 10−04 |
| GO-BP:0006796* | Phosphate-containing compound metabolic process | 24 | 1.44 × 10−04 |
| GO-BP:0006955 | Immune response | 15 | 7.26 × 10−03 |
| GO-BP:0006464* | Cellular protein modification process | 26 | 8.34 × 10−03 |
| GO-BP:0048731† | System development | 24 | 3.36 × 10−02 |
Enriched terms are symbol-coded to reflect relatedness in the ontology or functional proximity. *metabolic process; †developmental process; ‡response to stimulus. For each term, gene count shows number of genes in CDRs.
Domestication genes
To identify the key genes that play an important role in shaping the domestication of Enshi black pigs, we performed statistical analysis (Student’s t test) using the identity score (IS) of the two conditions (IS between the Enshi black pigs and the Chinese wild boar breeds or IS among three Enshi black pig populations). We then selected top 10 genes (ITPR3, RPS10, ERF, INSL6, ENSSSCG00000014230, TBX19, SFT2D2, VASN, GSK3A, and ORMDL1) with the lowest P value for further study.
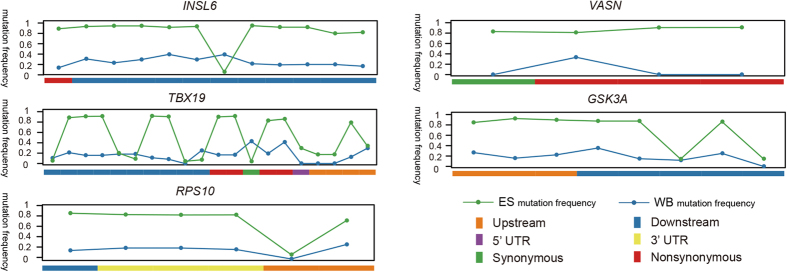
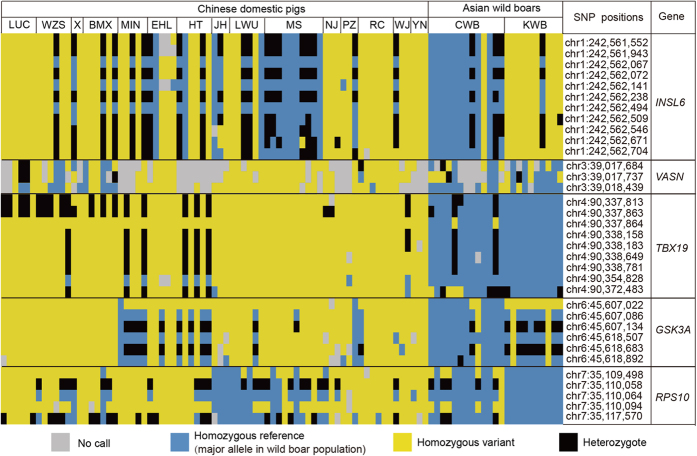
To analyze the signs of selection in detail, we detected SNPs from the regions spanning these genes with highly significant effects (SNPs located in untranslated regions (UTRs), exon, and downstream/upstream of the gene). We found 34 SNPs with significantly different mutation frequency between Enshi black pigs and Chinese wild boars (Fig. 5) in RPS10, GSK3A, INSL6, TBX19, and VASN. These SNPs may affect protein coding, gene splicing, transcription factor binding, and regulation of gene expression directly or indirectly. Furthermore, we genotyped the 34 SNPs in 15 pig breeds that represent a wide range of Chinese domestic pig populations (73 individuals), 13 Chinese wild boars, and 10 Korean wild boars11 (Fig. 6). The results demonstrated strong signatures of selection at these loci across Chinese domestic pigs that are used for pork production (i.e. muscle growth and adipose deposition). By combining the mutation frequency and genotyping results, we found that five genes were extremely different between the domestic pig breeds and Chinese wild boars. These genes may be responsible for the marked phenotypic changes produced by domestication of Enshi black pigs.
Figure 5. The difference of mutation frequency between Enshi black pig and Chinese wild boar.
The mutation frequency in Enshi black pig is the average of three Enshi populations.
Figure 6. The genotyping results of Asian wild boars and a diverse panel of Chinese domestic pigs.
Results from genotyping Asian wild boars and a diverse panel of Chinese domestic pigs for SNPs in the putative selective sweeps containing INSL6, VASN, TBX19, GSK3A, and RPS10. The plot is sorted by breeds but has not been clustered by individuals or by breeds. LUC, Luchuan; WZS, Wuzhishan; X, Xiang; BMX, Bamaxiang; MIN, Min; EHL, Erhualian; HT, Hetao; JH, Jinhua; LWU, Laiwu; MS, Meishan; NJ, Neijiang; PZ, Penzhou; RC, Rongchang; WJ, Wujin; YN, Yanan; CWB, Chinese wild boar; KWB, Korean wild boar.
Among these five genes, two genes may be associated with body size and immunity. The first gene is ribosomal protein S10 (RPS10), which harbors three mutations in the 3′ UTR, and has also been reported to be commonly mutated in Diamond-Blackfan Anemia16,17. This gene provides instructions for producing ~80 different ribosomal proteins, which are vital components of cellular structures called ribosomes18. RPS10 may be associated with body height, body length, and longissimus muscle weight in pigs19,20. Interestingly, a previous genome-wide association study (GWAS) also showed that RPS10 may be associated with limb bone length (which is associated with body height and body length)21. Our study confirmed this conclusion. The second gene is Vasorin (VASN), which has three nonsynonymous mutations. This gene may be involved in modulating arterial response to injury by inhibiting the TGF-β signaling pathway22,23,24. VASN is highly expressed in vascular smooth muscle cells (hence the name) and the developing skeletal system25. This expression pattern indicates that VASN may indirectly influence the body size of pigs during embryonic development.
As a typical meat-lard pig breed, Enshi pigs exhibit strong selection signals in the obesity-related gene glycogen synthase kinase 3 (GSK3). GSK3 is a constitutively active, proline-directed serine/threonine kinase that participates in a number of physiological processes that range from glycogen metabolism to gene transcription26. The gene exists in two isoforms, GSK3A and GSK3B27. GSK3A knockout experiment in mice displayed improved glucose tolerance in response to glucose load and elevated hepatic glycogen storage and insulin sensitivity28, which may result in obesity29,30. Association analysis revealed that the GSK3A Hin1I and MspI polymorphisms were significantly associated with loin muscle area, average back fat thickness, thorax-waist fat thickness, and buttock fat thickness31. In addition, the gene could influence the muscle-to-meat process via glycolysis, reduced pH, and pale color32,33. The selection of the GSK3 gene may contribute to enhanced fat storage ability and relatively high intramuscular adipose content.
The remaining two genes were involved in developmental processes and male fertility. TBX19 (known as TPIT) is a member of the T-box family of transcription factors, which are involved in the regulation of developmental processes34. Numerous reports demonstrated that mutation in this gene may isolate deficiency of pituitary POMC-derived ACTH35,36,37,38. ACTH deficiency is characterized by adrenal insufficiency symptoms, such as weight loss, lack of appetite, obesity, hypocortisolism, and low blood pressure39,40. Nonsynonymous mutations of this gene may exist in Tongcheng pigs3, and a strong selection signal was detected in all Chinese native pig in our analysis. These findings suggested that the selection of this gene exerted a comprehensive influence on all Chinese indigenous pigs. The final gene, insulin-like 6 (INSL6), is a member of the insulin superfamily, which comprises a group of structurally related proteins41. INSL6 is predominantly expressed in the testis, and it is necessary for the progression of spermatogenesis. Deficiency in INSL6 will cause varying levels of male infertility42,43,44,45,46.
These positively selected genes may serve as important genetic foundation of the evolutionary scenarios triggered by artificial selection for agricultural production of Enshi black pigs. Further studies are required to define the signaling cascades associated with these genes and their regulatory mechanisms in order to facilitate a better understanding of their roles in the formation of economically important native breeds.
Conclusions
This study detected the genomic signatures that may have shaped the domestication of Enshi black pigs in China. The genes found to be positively selected in Enshi pigs are involved in crucial biological processes such as body size and immunity (RPS10 and VASN), obesity (GSK3), male fertility (INSL6), and early development (TBX19). In addition, important mutations within these genes were also identified to enrich the pool of markers that can be used to further refine selection in these pigs in the future. Our research methods and findings should also be helpful in deciphering genomic footprints left by selection and domestication in other livestocks47,48,49.
Materials and Methods
Ethics statement
Animals care and all the experimentation in this study were carried out in accordance with the pre-approved guidelines from Regulation Proclamation No.5 of the Standing Committee of Hubei People’s Congress, and all experimental procedures and sample collection methods were approved by the Institutional Animal Care and Use Committee of Huazhong Agricultural University, Wuhan, China (permit HZAUSW2015-0003).
Animals and tissue collection
Three representative populations of Enshi black pig were raised in counties of Enshi Tujia and Miao Autonomous Prefecture in Hubei Province, China (LCP, YTL, and ZBZ; Fig. 1). Blood samples were obtained from 75 female individuals (31 from the LCP, 32 from the YTL, and 12 from the ZBZ).
Sequencing data
For each population, DNA samples from 12–32 individuals were pooled in equimolar quantities that were used to construct pair-end sequencing libraries with an insert size of 300 bp. The libraries were sequenced by Illumina Hiseq 4000 (Illumina, San Diego, CA, USA) with 150 bp paired-end reads according to the manufacturer’s instructions. We also downloaded 96 publicly available pig genomes in Asia (Supplementary Table S1), of which 13 Chinese wild boars were used to test for differentiation and possibly selection, and 10 Korean wild boars and 73 Chinese domestic pigs were used to investigate the patterns of selected loci.
To avoid reads with low-quality, we removed the following types of reads: (a) reads with ≥10% unidentified nucleotides (N); (b) reads with >10 nt aligned to the adapter, allowing ≤10% mismatches; and (c) reads with >50% bases having phred quality <5; and (d) putative PCR duplicates generated by PCR amplification in the library construction process. After the low-quality reads were excluded, the remaining high-quality reads were aligned against the Sscrofa10.2 reference sequence by using Burrows-Wheeler Aligner (BWA, v0.7.8), with the BWA command of “mem -t 4 -k 19 -M -w 200”. The uniquely aligned reads with ≤5 mismatches were used for SNP calling.
SNP calling and variation annotation
To obtain highly confident SNPs, we employed both SAMtools (v0.1.19) and GATK tool (v3.3) variant calling pipelines to process each pool of samples respectively. In SAMtools, base calling was conducted by using the “mpileup” command and the “-q 1 -C 50 -S -D -m 2 -F 0.002” parameters of SAMtools. The “view” command of BCFtools was used to convert the BCF files to VCF files. The VCF files were then filtered by the “vcfutils.pl” script with the use of the “varFilter -Q 20 -d 4 -D 1000” option and vcftools using the “–thin 4” option, and high-quality SNPs (coverage depth ≥4 and ≤1000, RMS mapping quality >20, the distance between adjacent SNPs ≥5 bp) were retained for subsequent analysis. For GATK, the settings were used according to the GATK best practice online documentation. Results that were obtained by the two pipelines were compared using the BEDTools (v2.21.0) “intersectBed” module50, and only concordant variations were processed further.
To determine novel variants in our sequence data, we compared the identified SNPs with the dbSNP (Build 143) data using BEDTools and annotated the detected genetic variants using ANNOVAR51. Gene ontology analysis was performed by the web-based software PANTHER52. Functional interaction network of CSGs was performed by the Reactome FI Plugin in the Cytoscape software environment53.
Selective-sweep analysis
For each population, we used allele counts at variable sites to identify signals of selection in 100 kb windows (with a step size of 50 kb) through two approaches: for each window, we calculated (1) the average pooled heterozygosity, HP , and (2) the average fixation index, FST, between Enshi black pig and Chinese wild boar. We calculated FST using PoPoolation254 and HP as follows:
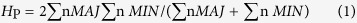 |
where nMAJ is the most frequently observed allele, and nMAJ is the least frequently observed allele.
Putatively selected regions in each group were located by extracting windows from the tails of the Z-transformed HP and FST distributions by applying a threshold of two standard deviations (P ≤ 0.05, Z(HP) < −2, Z(FST) > 2). We disregarded the selection signals on chromosome X because of the extremely low rate of recombination55 and ancient interspecies introgression7.
Calculation of identity score (IS)
We calculated ISs to visualize haplotype sharing in pairwise comparisons at the selected genes. For each identified SNP, we determined the fraction of reads that corresponded to the reference genome allele, F, in each pig population. The IS values of individual SNPs were then calculated as IS = 1 − (|FPopulation1 − FPopulation2|) (2), with SNPs assessed only if a minimum of one read was obtained in each population. The IS value for a gene was the mean of all IS values observed in the gene for a specific comparison.
Sequencing of the complete mitochondrial DNA (mtDNA) sequences
Ear tissue samples of five individuals were collected from the three counties (1 from LCP, 2 from YTL, and 2 from ZBZ; Supplementary Figure S4). DNA was isolated using a MicroElute Genomic DNA kit (OMEGA, USA). Nineteen pairs of primers (Supplementary Table S5) were used to amplify and sequence the complete mitochondrial DNA sequences. PCR reactions were performed using LA taq (TaKaRa, Dalian, China). PCR products were purified following agarose gel electrophoresis and then sequenced using the ABI 3730 DNA sequencer (Applied Biosystems, Foster City, CA, USA). Sequences of other representative domestic pigs and wild boars were downloaded from GenBank. The detailed information on the pig breeds are shown in Supplementary Table S6.
Phylogenetic analysis
To explore the genetic relationship of the three populations of Enshi black pigs and other pig breeds, we used the five newly generated complete mtDNA sequences, together with 18 downloaded complete mitochondrial DNA sequences, to perform phylogenetic analysis; the neighbor-joining tree was constructed using MEGA (v5.0)56.
To explore the genetic relationship of the 4 pooled group, we also constructed another two phylogenetic trees by using SNPs in whole genome and SNPs in selected regions respectively. Homologous regions among different pig populations were identified and extracted by SNPhylo (v20160204)57. The corresponding SNPs in homologous regions were utilized to construct a neighbor-joining tree using MEGA.
Additional Information
Accession codes: The genome resequencing reads have been deposited into the NCBI sequence read archive (SRA) under the accession SRP071318.
How to cite this article: Fu, Y. et al. Genomic analysis reveals selection in Chinese native black pig. Sci. Rep. 6, 36354; doi: 10.1038/srep36354 (2016).
Publisher’s note: Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary Material
Acknowledgments
We thank Professor Yaosheng Chen, Delin Mo, Ruiyi Lin and Cheng Zou for collecting pig samples for genome re-sequencing. This work was supported by a grant from the National Basic Research Priorities Program of China (973 Program, 2013CB835205), National Natural Science Foundation of China (31472076, 31522055, and 31472081), the Research Project of Chinese Ministry of Education (113048A) and the Program for Innovative Research Team of Sichuan Province (2015TD0012).
Footnotes
Author Contributions C.L. and M.L. conceived and designed the experiments and the analytical strategy. C.L. and J.L. performed animal work and prepared biological samples. Y.F., Q.T. and S.T. designed the bioinformatics analysis process. Y.F. and C.L. wrote the paper. M. L., C.L. and J.C. revised the paper. All authors reviewed the manuscript.
References
- Li M. et al. Whole-genome sequencing of Berkshire (European native pig) provides insights into its origin and domestication. Sci. Rep. 4, 4678 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rubin C. J. et al. Strong signatures of selection in the domestic pig genome. Proc. Natl. Acad. Sci. USA 109, 19529–19536 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang C. et al. Genome-wide analysis reveals artificial selection on coat colour and reproductive traits in Chinese domestic pigs. Mol. Ecol. Resour. 15, 414–424 (2015). [DOI] [PubMed] [Google Scholar]
- Wilkinson S. et al. Signatures of diversifying selection in European pig breeds. PLoS Genet. 9, e1003453 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang L. Animal Genetic Resources in China: Pigs (eds Wang L. et al.) (China Agriculture Press Beijing, 2011).
- Dai G. Industry Development and Genetic Resources Protection of Animal Husbandry in the World Selenium Capital Enshi Prefecture. Animal Husbandry and Feed Science 6, 144 (2014). [Google Scholar]
- Ai H. et al. Adaptation and possible ancient interspecies introgression in pigs identified by whole-genome sequencing. Nat. Genet. 47, 217–225 (2015). [DOI] [PubMed] [Google Scholar]
- Frantz L. A. et al. Evidence of long-term gene flow and selection during domestication from analyses of Eurasian wild and domestic pig genomes. Nat. Genet. 47, 1141–1148 (2015). [DOI] [PubMed] [Google Scholar]
- Groenen M. A. et al. Analyses of pig genomes provide insight into porcine demography and evolution. Nature 491, 393–398 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li M. et al. Genomic analyses identify distinct patterns of selection in domesticated pigs and Tibetan wild boars. Nat. Genet. 45, 1431–1438 (2013). [DOI] [PubMed] [Google Scholar]
- Moon S. et al. A genome-wide scan for signatures of directional selection in domesticated pigs. BMC Genomics 16, 130 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li H. & Durbin R. Fast and accurate long-read alignment with Burrows-Wheeler transform. Bioinformatics 26, 589–595 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li H. et al. The Sequence Alignment/Map format and SAMtools. Bioinformatics 25, 2078–2079 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- McKenna A. et al. The Genome Analysis Toolkit: a MapReduce framework for analyzing next-generation DNA sequencing data. Genome Res. 20, 1297–1303 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jeong H. et al. Exploring evidence of positive selection reveals genetic basis of meat quality traits in Berkshire pigs through whole genome sequencing. BMC Genet. 16, 104 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boria I. et al. The ribosomal basis of Diamond-Blackfan Anemia: mutation and database update. Hum. Mutat. 31, 1269–1279 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Doherty L. et al. Ribosomal protein genes RPS10 and RPS26 are commonly mutated in Diamond-Blackfan anemia. Am. J. Hum. Genet. 86, 222–228 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lindstrom M. S. Emerging functions of ribosomal proteins in gene-specific transcription and translation. Biochem. Biophys. Res. Commun. 379, 167–170 (2009). [DOI] [PubMed] [Google Scholar]
- Wang L. et al. Genome-wide association studies identify the loci for 5 exterior traits in a Large White x Minzhu pig population. PLoS One 9, e103766 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao S. et al. Functional analysis of inter-individual transcriptome differential expression in pig longissimus muscle. J. Anim. Breed Genet. 130, 72–78 (2013). [DOI] [PubMed] [Google Scholar]
- Zhang L. C. et al. A genome-wide association study of limb bone length using a Large White x Minzhu intercross population. Genet. Sel. Evol. 46, 56 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang A. et al. Exosomal transfer of vasorin expressed in hepatocellular carcinoma cells promotes migration of human umbilical vein endothelial cells. Int. J. Biol. Sci. 11, 961–969 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li S. et al. Vasorin is a potential serum biomarker and drug target of hepatocarcinoma screened by subtractive-EMSA-SELEX to clinic patient serum. Oncotarget 6, 10045–10059 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Malapeira F. et al. Cancer degradomics: ADAMI7 regulates TGF-BETA signaling through the cleavage of vasorin (2011).
- Krautzberger A. M., Kosiol B., Scholze M. & Schrewe H. Expression of vasorin (Vasn) during embryonic development of the mouse. Gene Expr. Patterns 12, 167–171 (2012). [DOI] [PubMed] [Google Scholar]
- Hooper C., Killick R. & Lovestone S. The GSK3 hypothesis of Alzheimer’s disease. J. Neurochem. 104, 1433–1439 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bhattacharjee R. et al. Targeted disruption of glycogen synthase kinase 3A (GSK3A) in mice affects sperm motility resulting in male infertility. Biol. Reprod. 92, 65 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- MacAulay K. et al. Glycogen synthase kinase 3alpha-specific regulation of murine hepatic glycogen metabolism. Cell Metab. 6, 329–337 (2007). [DOI] [PubMed] [Google Scholar]
- Mohsin F., Mahbuba S., Begum T., Azad K. & Nahar N. Prevalence of impaired glucose tolerance among children and adolescents with obesity. Mymensingh Med. J. 21, 684–690 (2012). [PubMed] [Google Scholar]
- Muscelli E. et al. Separate impact of obesity and glucose tolerance on the incretin effect in normal subjects and type 2 diabetic patients. Diabetes 57, 1340–1348 (2008). [DOI] [PubMed] [Google Scholar]
- Wang L. et al. Identification of porcine glycogen synthase kinase 3alpha (GSK-3alpha) gene and its association with carcass traits. Mol. Cell Biochem. 377, 65–73 (2013). [DOI] [PubMed] [Google Scholar]
- Bowker B., Grant A., Forrest J. & Gerrard D. Muscle metabolism and PSE pork. Journal of Animal Science 79, 1–8 (2000). [Google Scholar]
- Liu X. et al. Muscle Transcriptional Profile Based on Muscle Fiber, Mitochondrial Respiratory Activity, and Metabolic Enzymes. Int. J. Biol. Sci. 11, 1348–1362 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yi C. H. et al. Identification, mapping, and phylogenomic analysis of four new human members of the T-box gene family: EOMES, TBX6, TBX18, and TBX19. Genomics 55, 10–20 (1999). [DOI] [PubMed] [Google Scholar]
- Eriksson K. S. & Mignot E. T-box 3 is expressed in the adult mouse hypothalamus and medulla. Brain Res. 1302, 233–239 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kojima T. & Takahashi N. Hox genes and teratogenic factors (INTECH Open Access Publisher, 2012). [Google Scholar]
- Liu J. et al. Tbx19, a tissue-selective regulator of POMC gene expression. Proc. Natl. Acad. Sci. USA 98, 8674–8679 (2001). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wagner J. & Thomas P. Genetic determinants of mammalian pituitary morphogenesis. Front. Biosci. 12, 125–134 (2007). [DOI] [PubMed] [Google Scholar]
- Mendiratta M. S. et al. Early onset obesity and adrenal insufficiency associated with a homozygous POMC mutation. Int. J. Pediatr. Endocrinol. 2011, 5 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pulichino A. M. et al. Human and mouse TPIT gene mutations cause early onset pituitary ACTH deficiency. Genes. Dev. 17, 711–716 (2003). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brailoiu G. C. et al. Insulin-like 6 immunoreactivity in the mouse brain and testis. Brain Res. 1040, 187–190 (2005). [DOI] [PubMed] [Google Scholar]
- Chan L. J., Hossain M. A., Samuel C. S., Separovic F. & Wade J. D. The relaxin peptide family–structure, function and clinical applications. Protein Pept. Lett. 18, 220–229 (2011). [DOI] [PubMed] [Google Scholar]
- Ivell R. & Grutzner F. Evolution and male fertility: lessons from the insulin-like factor 6 gene (Insl6). Endocrinology 150, 3986–3990 (2009). [DOI] [PubMed] [Google Scholar]
- Lok S. et al. Identification of INSL6, a new member of the insulin family that is expressed in the testis of the human and rat. Biol. Reprod. 62, 1593–1599 (2000). [DOI] [PubMed] [Google Scholar]
- Lu C., Lam H. N. & Menon R. K. New Members of the Insulin Family: Regulators of Metabolism, Growth and Now … Reproduction. Pediatric Research 57, 70R–73R (2005). [DOI] [PubMed] [Google Scholar]
- Verbeken M., Wynendaele E., Lefebvre R. A., Goossens E. & Spiegeleer B. D. The influence of peptide impurity profiles on functional tissue-organ bath response: the 11-mer peptide INSL6[151-161] case. Anal. Biochem. 421, 547–555 (2012). [DOI] [PubMed] [Google Scholar]
- Boitard S., Boussaha M., Capitan A., Rocha D. & Servin B. Uncovering Adaptation from Sequence Data: Lessons from Genome Resequencing of Four Cattle Breeds. Genetics 203, 433–450 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guo X. et al. Whole-genome resequencing of Xishuangbanna fighting chicken to identify signatures of selection. Genet Sel Evol 48, 62 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang J. et al. Whole-Genome Sequencing of Native Sheep Provides Insights into Rapid Adaptations to Extreme Environments. Mol Biol Evol 33, 2576–2592 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Quinlan A. R. & Hall I. M. BEDTools: a flexible suite of utilities for comparing genomic features. Bioinformatics 26, 841–842 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang K., Li M. & Hakonarson H. ANNOVAR: functional annotation of genetic variants from high-throughput sequencing data. Nucleic Acids Res. 38, e164 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mi H., Poudel S., Muruganujan A., Casagrande J. T. & Thomas P. D. PANTHER version 10: expanded protein families and functions, and analysis tools. Nucleic Acids Res. 44, D336–D342 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shannon P. et al. Cytoscape: a software environment for integrated models of biomolecular interaction networks. Genome Res. 13, 2498–2504 (2003). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kofler R., Pandey R. V. & Schlotterer C. PoPoolation2: identifying differentiation between populations using sequencing of pooled DNA samples (Pool-Seq). Bioinformatics 27, 3435–3436 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ma J. et al. Recombinational landscape of porcine X chromosome and individual variation in female meiotic recombination associated with haplotypes of Chinese pigs. BMC Genomics 11, 159 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tamura K. et al. MEGA5: molecular evolutionary genetics analysis using maximum likelihood, evolutionary distance, and maximum parsimony methods. Mol. Biol. Evol. 28, 2731–2739 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee T. H., Guo H., Wang X., Kim C. & Paterson A. H. SNPhylo: a pipeline to construct a phylogenetic tree from huge SNP data. BMC Genomics 15, 162 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.