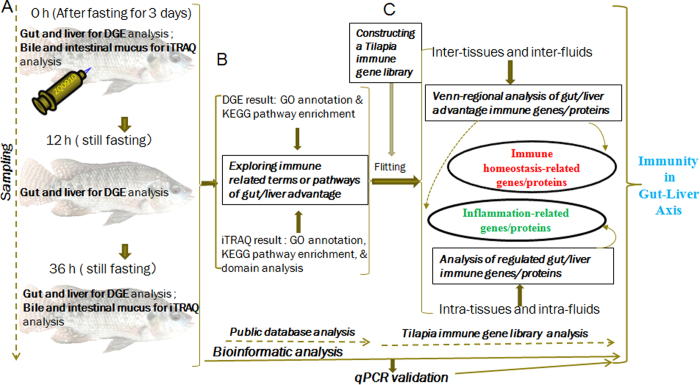
Figure 1. Strategy for identification of regulated immune genes in fish gut and liver by integrative analysis of both DGE transcriptomic and iTRAQ-based quantitative proteomic data.
(A) Tilapias were infected intraperitoneally with live S. agalactiae, and then sampling for DGE profiling of gut or liver was done at 0 h (just before challenge), together with 12 h and 36 h post challenge, meanwhile sampling for iTRAQ analysis of intestinal mucus or bile was done at 0 h and 36 h, from both healthy and infected fish; (B) Immune related annotation was found out via bioinformation analysis of both DGE and iTRAQ results using public database (including GO, KEGG, and InterPro); (C) On one hand, the comparison was done in both inter-tissue/fluid data (gut vs liver, or intestinal mucus vs bile) and intra-tissue/fluid data (gut, liver, mucus or bile), then Venn-regional analysis was applied to screened out immune homeostasis-related transcripts and proteins; on the other hand, regulated gut-liver immune transcripts or proteins were revealed by intra-tissue (same tissue at different time-points) or intra-fluid (same fluid at different time-points) comparison.