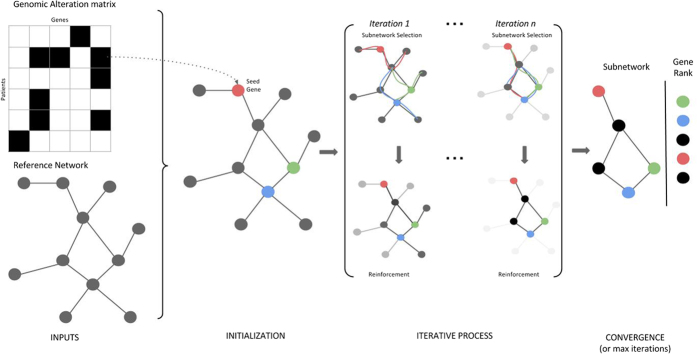
Figure 1. Overview of SSA-ME.
The input consists of a matrix containing genomic alterations (i.e. mutations or copy number alterations, among others) across patients (depicted as black tiles) and a human reference network. In a first initialization step, every gene which has at least one genomic alteration across all patients is selected as a seed gene (colored genes in the network). The gene scores (represented as the opacity of the genes in the networks) are uniformly set to a value of 0.5. In every subsequent iteration step, small subnetworks will be generated, starting at every seed gene. Every gene adjacent to the small subnetwork has a chance proportional to its score to be incorporated in the small subnetwork. When a certain size has been reached the small subnetwork generation will stop and a score for each selected small subnetwork will be calculated based on the mutually exclusivity pattern found within this small subnetwork. At the end of every iteration step these small subnetwork scores will be used to update gene scores, altering the chance of genes to be incorporated into the small subnetwork in subsequent iteration steps. Upon convergence it can be seen that a few genes have high scores while others have scores close to 0. Genes are ranked based on their gene scores which reflects their potential to belong to a mutual exclusivity pattern.