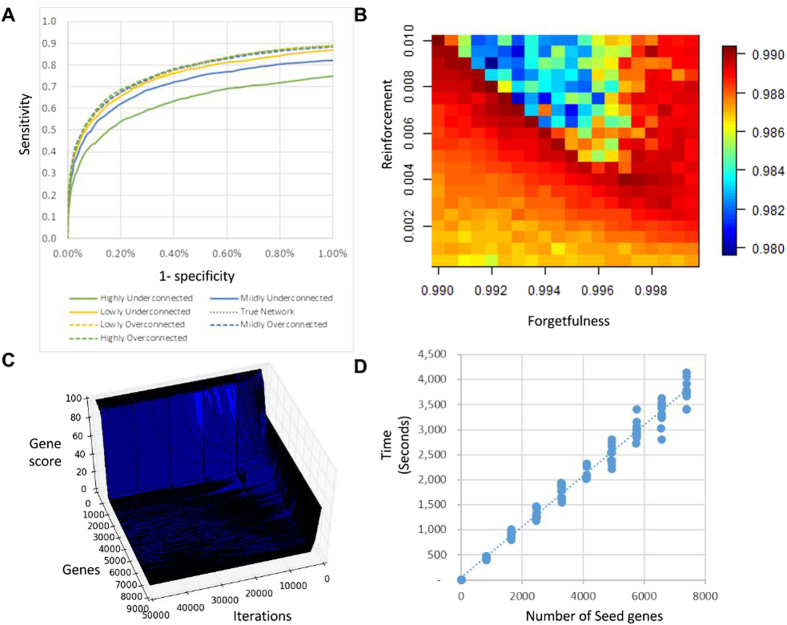
Figure 2. Performance on Simulated Data.
(A) Robustness of the predictions with respect to the used reference network. The X-axis represents 1-specificity and the Y-axis represents sensitivity (ROC curve). Underconnected networks lead to lower performance while overconnected networks result in similar, although lower, performance as compared to the performances obtained with the original network. Note that, for clarity reasons, the range of the x-axis is restricted to [0, 0.01]. (B) Heat map depicting parameter sensitivity. Area under the ROC curve (AUC) values for every analyzed parameter pair are depicted. Warm colors depict higher AUC values while cold colors depict lower AUC values. It can be seen that the best performance is achieved on the diagonal for combinations of reinforcement and forgetfulness of 1. (C) Plot visualizing convergence and stability of convergence of gene scores. The X-axis represents the number of performed iterations, the Y-axis displays all genes in the reference network (black lines in the plot) and the Z-axis represents the gene scores. All genes start on the right side with a gene score of 0.5. Most of them converge fast to 0 or 1. As no inflecting lines are observed, convergence is stable. Results are shown on a plot depicting scores for all genes at every iteration step. (D) Plot showing linear time complexity of the algorithm with respect to the number of seed genes. Each dot on the plot represents the time to convergence of a separate run. Per tested number of seed genes, 10 simulations were performed. Results were obtained by running the algorithm on one single processor Intel(R) Xeon(R) CPU E5-2670 0 @ 2.60 GHz.