Fig. 2.
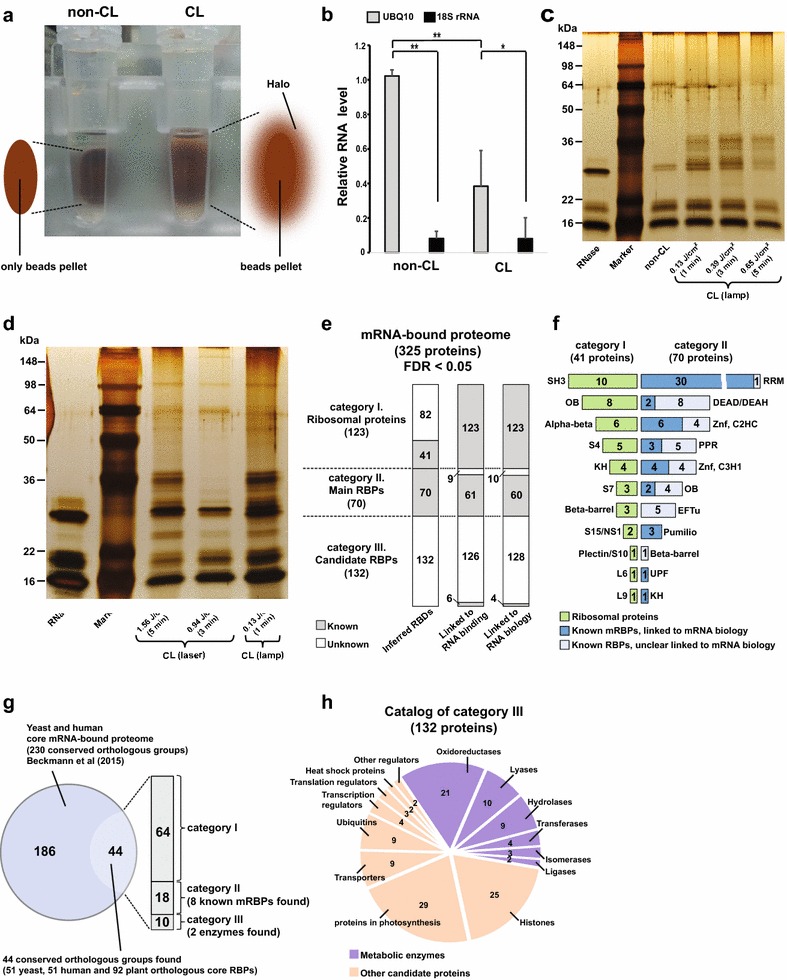
mRNA-bound proteome from Arabidopsis leaf mesophyll protoplasts. Observed halo surrounding beads pellet in CL sample and not in non-CL sample during wash step (a). 18S rRNA and UBQ10 mRNA expression levels in non-CL and CL samples by qRT-PCR (values were mean ± SD (n = 3); single asterisk and double asterisk significant differences with p < 0.05 and <0.01) (b). Separated mRBPs in protein eluent by SDS-PAGE gels and visualized by silver-staining (c, d). Protein eluent of non-CL sample compared with CL samples irradiated by continuous UV for 1, 3 and 5 min (c). CL sample irradiated by 1 min continuous UV compared with CL samples irradiated by a pulsed UV laser source for 3 and 5 min (d). Classification of three categories from mRNA-bound proteome (quantity of identified proteins listed in numbers and the false discovery rate (FDR) at the peptide and protein levels below 5%) (e). List of proteins from category I and II according to the annotated RNA-binding domains (f). Detection of plant orthologous core RBPs to yeast and human through comparison between our mRNA-bound proteome and the core mRNA-bound proteome of yeast and human from literature Beckmann et al. [16] (g). Pie chart of classification of category III. Quantity of identified metabolic enzymes and other candidate proteins listed in numbers (h)
