Summary
Corticothalamic projection neurons (CThPN) are a diverse set of neurons, critical for function of the neocortex. CThPN development and diversity needs to be precisely regulated, but little is known about molecular controls over their differentiation and functional specialization, critically limiting understanding of cortical development and complexity. We report the identification of a set of genes that both define CThPN, and likely control their differentiation, diversity, and function. We selected the CThPN-specific transcriptional co-regulator Fog2 for functional analysis. We identify that Fog2 controls CThPN molecular differentiation, axonal targeting, and diversity, in part by regulating the expression level of Ctip2 by CThPN, via combinatorial interactions with other molecular controls. Loss of Fog2 specifically disrupts differentiation of subsets of CThPN specialized in motor function, indicating that Fog2 coordinates subtype and functional- area differentiation. These results confirm that we identified key controls over CThPN development, and identify Fog2 as a critical control over CThPN diversity.
Introduction
Corticothalamic projection neurons (CThPN) are critical for the function of the cerebral cortex. CThPN broadly control access of sensory information to the cortex by modulating the activity of the thalamus, and locally regulate the activation state of other type of neurons across cortical layers in their area (Bortone et al., 2014; Olsen et al., 2012). CThPN are quite diverse. Most CThPN reside in layer VI, while a small proportion resides in layer V. In distinct cortical areas, layers, and sublaminae, they are specialized to process distinct modalities of information (Briggs et al., 2010). Abnormal CThPN specification, differentiation, and connectivity would be expected to disrupt their functional integration, and therefore critically impact cortical function. Surprisingly, little is known about molecular mechanisms that control CThPN development and diversity. More broadly, mechanisms that govern the emergence of neuronal diversity in the neocortex are beginning to be elucidated, and still are not well understood. Because of their functional specialization and diversity, CThPN development provides a particularly informative model for investigation of these broader issues of neuronal diversity in the neocortex.
Differentiation of distinct types of neurons requires execution of elaborated developmental programs that progressively direct specification of progenitors into neurons with specific subtype and area identities. Cortical projection neurons are generated sequentially from pallial progenitors, migrate to birthdate-appropriate layers, and differentiate into broad subtypes of neurons, which are further specialized within distinct cortical areas, adopting characteristic gene expression and connectivity. Subplate neurons (SPN), CThPN, and subcerebral projection neurons (SCPN) are closely-related neuron subtypes that belong to the broader class of corticofugal projection neurons (CFuPN), which all project away from the cortex. They are sequentially generated at approximately embryonic days (E) 11.5, E12.5, and E13.5, and primarily populate the subplate, layer VI, and layer V, respectively. In contrast, callosal projection neurons (CPN) connect within the cortex to targets in the contralateral hemisphere. Most CPN are born around E15.5 and reside primarily in layers II-III (Greig et al., 2013).
Recently, substantial progress has been made toward understanding molecular mechanisms controlling projection neuron subtype development, identifying key controls over initial fate decisions during specification of broad classes of projection neurons, in particular, between CFuPN and/or SCPN versus CPN. Cross-repressive interactions between controls from alternative subtype programs, such Fezf2, Ctip2, Sox5, Tbr1, and Satb2, progressively define broad subtype identities (Arlotta et al., 2005; Chen et al., 2005a, Chen et al., 2005b; Molyneaux et al., 2005; Chen et al., 2008; Kwan et al., 2008; Lai et al., 2008; Bedogni et al., 2010; Han et al., 2011; McKenna et al., 2011; Srinivasan et al., 2012; Greig et al., 2013). However, beyond this initial fate specification, little is known about molecular programs underlying differentiation and diversity of CThPN, or any projection neuron subtype. Understanding mechanisms that generate the extraordinary diversity of neuron subtypes and sub-subtypes in the neocortex requires much deeper knowledge of neuron differentiation, beyond the specification of broad classes of projection neurons; this includes identifying controls acting at more advanced stages in differentiation that regulate the multiple variables that define the n-dimensional ‘identity space’ characteristic of distinct neuron subtypes, such as axonal projection, dendritic morphology, synaptic specificity, electrophysiological properties, etc., and that ultimately define neuron functional identity. Investigating controls regulating neuronal subtype differentiation at these distinct levels is critical for understanding neuronal development and diversity, since these controls are likely to more directly contribute to the diversification of broad neuronal classes of neurons into functional subsets (also relevant to function, evolution, regeneration, and optimal directed differentiation).
Previous work from our laboratory and others has identified molecular controls over development of corticospinal motor neurons (CSMN), CPN, and neocortical projection neurons more broadly (Arlotta et al., 2005; Molyneaux et al., 2009; Greig et al., 2013), but almost nothing is known about controls over CThPN development and diversity. To address this directly, we purified CThPN at 3 important developmental stages, and compared their gene expression with distinct neocortical projection neuron subtypes: CSMN, which also belong to the broader class of CFuPN; and CPN, which make starkly different axonal trajectories, projecting across the midline to contralateral cortex.
We first identified a novel set of genes progressively restricted to CThPN, with largely uncharacterized functions in cortical development. Some of these genes are expressed by all CThPN, throughout differentiation or at distinct developmental stages, some are restricted to specific CThPN subpopulations, and some display shared expression with other subtypes, typically SCPN. We next investigated one of the CThPN- specific genes as a candidate CThPN molecular control: Zfpm2, also known as ‘Friend of GATA-2’ (Fog2). Fog2 is a transcriptional co-regulator with important functions in cell differentiation in several tissues, including cardiac, gonadal, mammary gland, and lung (Tevosian et al., 2000, 2002; Ackerman et al., 2007; Manuylov et al., 2007), but its function in the nervous system was previously unknown.
Here, we report that Fog2 coordinates two aspects of CThPN development — subtype differentiation from other neuron types, and acquisition of area/functional identity— in part by regulating the level of CTIP2. In the absence of Fog2, CThPN in motor cortex express high level Ctip2, and their axonal projections to motor thalamus are disrupted. Mis-expression of Fog2 in SCPN reduces Ctip2 expression, leading to reduced projection to the cerebral peduncle. Fog2 function intersects combinatorially with additional molecular controls in an area-specific manner. This intersectional mechanism serves to increase neuronal diversity of CThPN. Beyond identifying Fog2 as a central CThPN developmental control, these studies identify what are likely other combinatorial molecular controls over distinct aspects of CThPN development, from broad subtype specification to more refined and areally-restricted features of CThPN identity and function.
Results
Identification of CThPN-Genes
To identify CThPN developmental controls, we first compared gene expression of purified CThPN at three important developmental stages with the gene expression of CSMN and CPN at the same ages, as previously described in Arlotta et al., 2005 and Molyneaux et al., 2009. Briefly, we retrogradely labeled CThPN via ultrasound guided microinjection of fluorescent microspheres into the developing thalamus, and isolated CThPN by FACS at E18, postnatal day 3 (P3), and P6. From purified CThPN, RNA was isolated, amplified, and hybridized on Affymetrix microarray chips. To identify CThPN- specific genes, statistical significance of gene expression differences between neuron subtypes was determined by pairwise comparisons at each age. Only genes with a minimum of a three-fold difference (p < 0.01) in expression level in relation to the comparator populations at any stage were chosen for further analysis. We selected the genes with the most significant CThPN-specific expression at any single time from each comparison for further study by Gene Ontology (GO) analysis, and by analysis of temporal dynamics of expression. All microarray data are available at the GEO database (Accession GSE61711).
CThPN-specific genes that we identified encode molecules that belong to multiple functional classes, and involved in diverse biological processes, including regulation of transcription (e.g., Fog2, Gas7, Nfia, Nfib, Phr1, Rnpc2, Shb, Tle4, Tbr1, Zmiz1); chromatin remodeling (e.g., Mllt1, Smarcd3, Zmyndll); axonal growth and guidance (e.g., Itsn1, Phr1, Sema5a); cell adhesion (e.g., Cdh9, Cntn2, Col5a1, Dlg5, Sdk1); synaptic signaling (e.g., Cacna1e, Drd1a, Gabrb3, Kcnk1, Kcnq2); and regulation of multiple important signaling pathways, including Notch, Wnt, and IGF, among others (e.g., Cxxc5, Igfbp3, Pcnx, Ror2). We performed GO-term enrichment analysis (GoProfiler and GoTerm-finder) to determine biological processes highly represented in this gene group. Sixty six GO-terms for biological processes are enriched (p<0.01, Benjamini-Hochberg false discovery rate FDR<5%). Some terms are too broad to interpret, such us ‘cell development’, but other terms are directly relevant to CThPN function (e.g. ‘response to stimulus’, ‘neuron projection morphogenesis’, ‘learning and memory’, or ‘synaptic transmission’; Table S1).
To identify top candidate genes of high biological relevance in CThPN development and/or function, we further applied filters focused on biological criteria, such as known functions of the genes in other systems, expression during CThPN development, and correlation of gene function and expression level with the unique developmental stages of CThPN and comparator neuron subtypes. We identified top candidate genes of potential high biological relevance (30 genes, Figure 1), which we classified as follows: (1) genes enriched in CThPN at all stages analyzed, which might be important for both establishment and maintenance of CThPN identity (Figure 1A); (2) genes enriched in CThPN early in development, which might be important for CThPN specification and early differentiation (Figure 1B); (3) genes for which expression peaks as CThPN develop, potentially controlling intermediate aspects of differentiation, such as axon outgrowth and targeting (Figure 1C); (4) genes enriched at later stages, which might control later events, such as synaptic targeting and maturation (Figures 1D); (5) genes excluded from CThPN, but expressed by other cortical projection neuron populations (Figure 1E).
Figure 1. CThPN-specific genes of high biological relevance for corticothalamic development classified based on temporal expression.
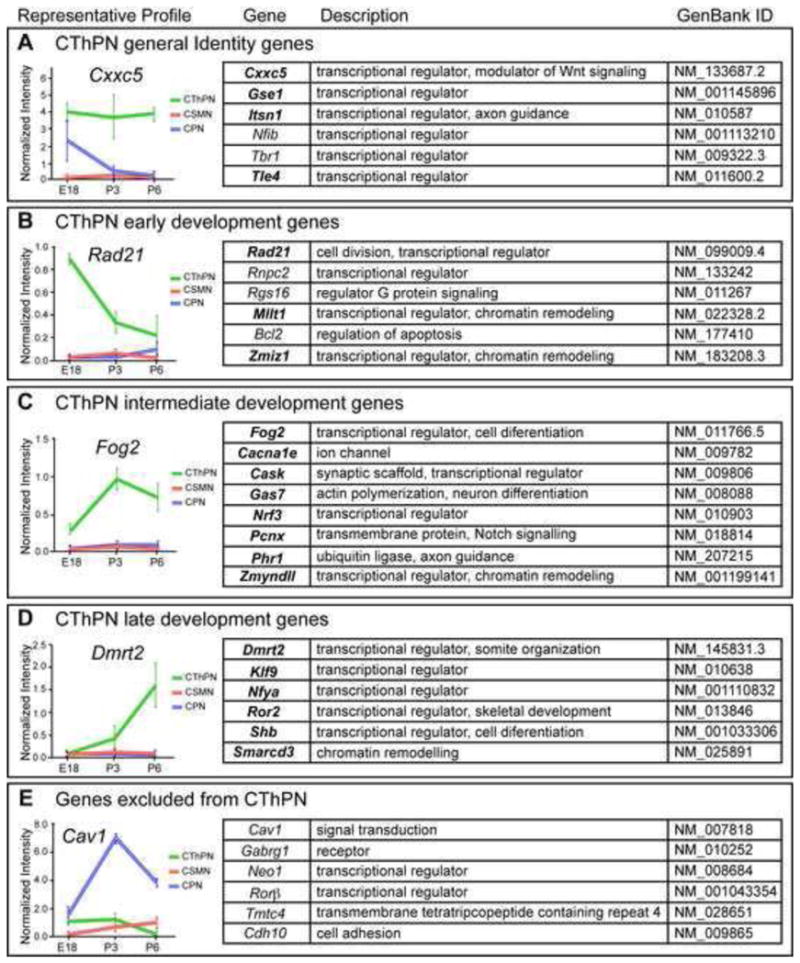
A set of biologically particularly relevant genes is shown, selected from the larger group of differentially expressed genes (available at GSE61711) based on function in other systems and temporal match to relevant biological processes by CThPN. Each group is represented by a prototypical expression profile shown on the left. (A) General Identity Genes: enriched in CThPN at all stages of development; (B) Early Development Genes: highly expressed by CThPN early in development; (C and D) Intermediate Development and Late Development Genes: exhibit increasing levels of expression as CThPN develop; and (E) Genes excluded from CThPN: expressed at high levels in other cortical projection neuron subtypes, serving as negative markers for CThPN. Expression by ISH and graphic expression profiles are shown for all genes listed in bold, either in Figure 2 or Figure S1. Data are represented as mean ± SEM.
We confirmed that genes frequently used as layer VI or CThPN markers, such as Tbr1, Tle4, Foxp2, and Nfib (Bedogni et al., 2010; Yao et al., 1998; Ferland et al., 2003; Betancourt et al., 2014), are present in our set of highly relevant candidate genes (Figure 1). Importantly, Tbr1 controls CThPN identity (Bedogni et al., 2010; McKenna et al., 2011; Han et al., 2011), and Nfib is necessary for CFuPN axon growth (Betancourt et al., 2014). These gene identifications both serve as positive internal controls, reinforcing the validity of this screen for CThPN subtype-specific developmental controls, and support the likely relevance of the much broader set identified.
We confirmed by in situ hybridization (ISH), immunocytochemistry (ICC), and exploration of expression databases (Genepaint, Allen Brain Atlas, BGEM) that each gene in this set is expressed in developing layer VI, where CThPN are the predominant neuronal subtype (Figure 2, Figure S1). We selected a representative subset of these genes to further investigate their expression over developmental time by ISH, confirming that the microarray results match the temporal expression identified by ISH (Figure 2).
Figure 2. Expression analysis of CThPN-specific genes by in situ hybridization parallels the temporal expression results from microarrays.
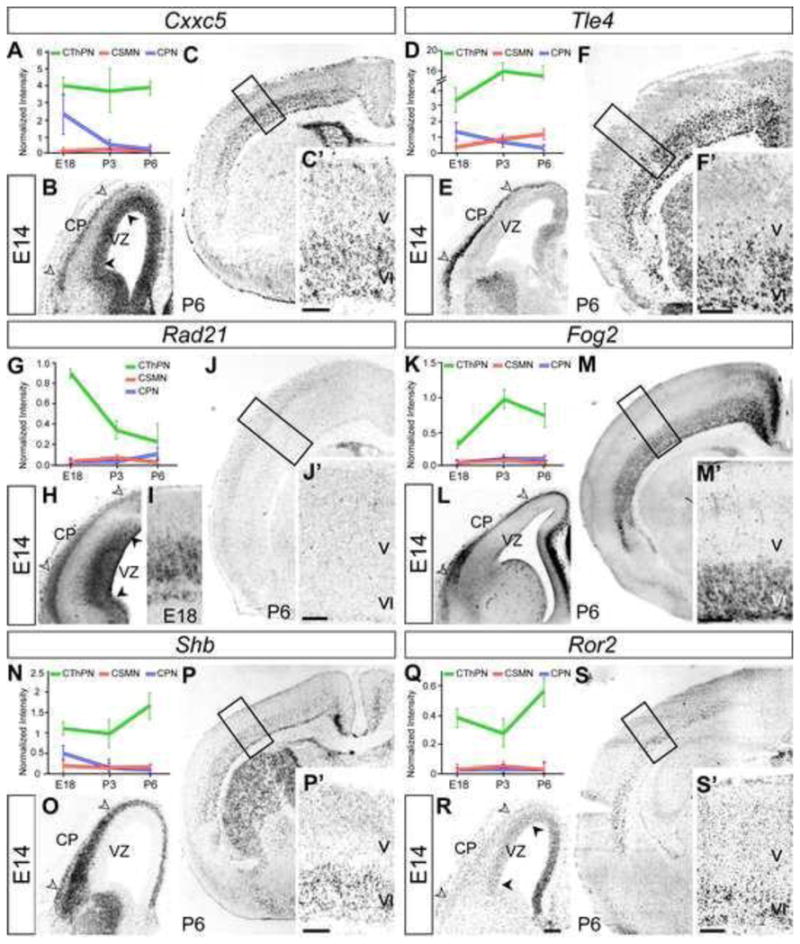
Expression analysis, including graphic expression profiles from microarray and ISH, of genes representing each category listed in Figure 1 (bold). Cxxc5 and Tle4 (A–F) are representative examples of “general identity genes”. Rad21 (G–J) and Fog2 (K–M) exemplify “early development” and “intermediate development” gene categories. Ror2 and Shb represent “late development genes” (N–S). (B, E, H, L, O, R) White arrowheads indicate expression in the cortical plate (CP), and black arrowheads indicate expression in the ventricular zone (VZ). Scale bars, 200 μm.
Genes identified have a range of specificity for CThPN (Figure 2, Figure S1). Some genes are highly expressed by CThPN, and are expressed in varying degrees by other subtypes, most often by neurons in layer V, suggesting that these genes are expressed by broader sets of CFuPN (e.g., Cacna1e, Foxp2, Nrf3, Tle4). Other subtype linkages also exist; e.g., we identified one gene expressed by CThPN and by neurons in layer IV (Klf9). We also identify genes expressed exclusively in layer VI across cortical areas (e.g., Fog2, Gse1, Itsn1, Gas7, Mllt1, Nfya, Rad21), likely by all CThPN (Figure 2, Figure S1). We identify yet other genes restricted to layer VI, but differentially expressed by CThPN subpopulations across distinct cortical areas. Some are expressed with spatial gradients; for example, Cxxc5, Ror2, Shb, Smarcd3, and Pcnx are expressed in medio-lateral or rostro-caudal gradients. Others have area-specific enrichment; Phr1, Zmiz1, and Zmynd11 are enriched in somatosensory cortex, while Dmrt2 is excluded from it. Still others subdivide layer VI into sublayers; Cask and Ror2 are highly expressed by CThPN located in the lower half of layer VI and subplate, and are weakly expressed by CThPN in upper layer VI (Figure 2, Figure S1).
Together, these experiments identify a novel set of genes expressed with a range of specificity by CThPN and CThPN subsets, mostly with uncharacterized functions in cortical development, and whose combinatorial expression progressively defines CThPN identity and diversity.
Fog2 is expressed by CThPN during postmitotic differentiation
From the highly relevant identified candidate CThPN controls, we selected Fog2 for first in-depth functional investigation. Fog2 is a non-DNA binding co-repressor that interacts with transcription factors such as Gata and Coup-tfs to regulate transcription (Tevosian et al., 2002; Huggins et al., 2001). Fog2 temporal expression, with maximal enrichment at intermediate stages of development (Figure 1C, Figure 2K), together with its predicted co-repressor function, makes it a compelling candidate to regulate intermediate temporal aspects of CThPN development, with strong potential for contributing to CThPN neuron class differentiation and diversification. Importantly, though high expression level in the brain was reported when Fog2 was first described (Tevosian et al., 1999), and though functions in development of the Drosophila nervous system have been suggested (Cubadda et al., 1997), no function in mammalian CNS development has been previously identified. In neocortex, Fog2 has been used as layer VI marker (Kwan et al., 2008; McKenna et al., 2011), but its function in cortical development has not been investigated.
We first confirmed the specificity of Fog2 to CThPN, and ruled out expression by other neuron subtypes. We retrogradely labeled CThPN, CSMN, and CPN with FluoroGold (FG), and performed ICC for FOG2. We identified that virtually all CThPN (FG labeled) express Fog2 (Figure 3A), and we confirmed that Fog2 is not expressed by CSMN or CPN (Figure 3 B–C). We further examined whether Fog2 is expressed by any SCPN or CPN by ICC for CTIP2 (highly expressed by SCPN) or SATB2 (expressed by CPN). Fog2 is not expressed by neurons with high level expression of Ctip2 (Figure S2 D–F). We also find that Satb2 expressing neurons, even those located in layer VI intermingled with CThPN, do not express Fog2, further supporting CThPN specificity (Figure S2 G–I). Since Tbr1 is frequently used as a CThPN marker, we compared the subtype specificity of Fog2 and Tbr1 expression. We find that, in addition to CThPN, Tbr1 is expressed by a number of CPN in layer VI, while Fog2 is exclusively expressed by CThPN (Figure S2 J–N).
Figure 3. Fog2 is expressed by postmitotic corticothalamic projection neurons, but is excluded from corticospinal and callosal projection neurons.
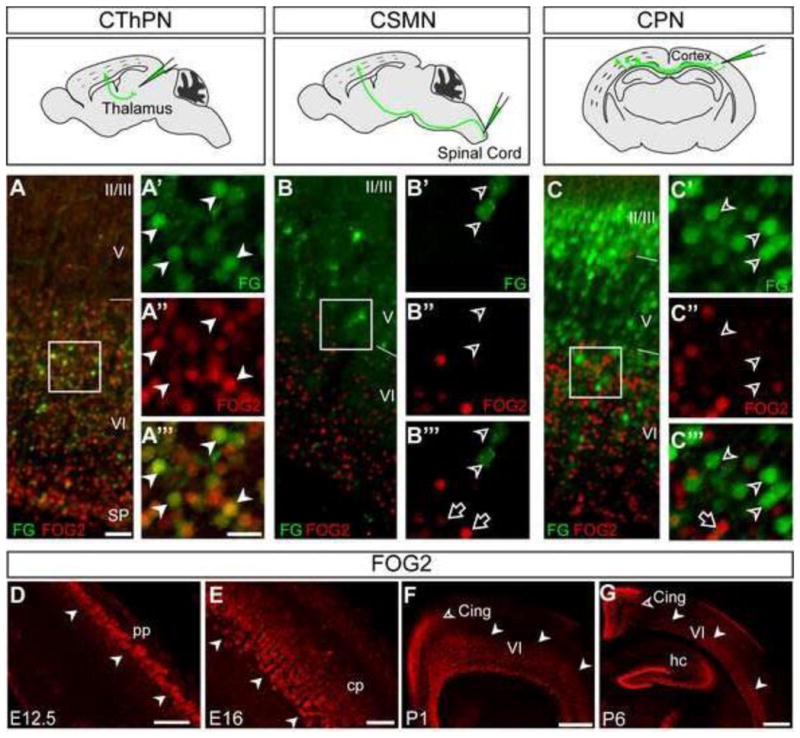
(A–C‴) FOG2 ICC (red) on coronal sections from brains injected with FluoroGold (FG) in thalamus, spinal cord, or contralateral cortex to retrogradely label CThPN, CSMN, or CPN, respectively (green). Schematic views of FG injection sites are at the top of each panel group. (A-A‴) FOG2 co-localizes with FG in retrograde labeled CThPN (white arrowheads in A′-A‴, area boxed in A). FG-labeled CSMN (B-B‴), or CPN (C-C‴) do not express FOG2. (D–G) Time-course of FOG2 expression revealed by ICC. FOG2 is first expressed at E12.5 in the preplate (white arrowheads) (D). FOG2 is strongly expressed in deep cortical plate during embryonic development (E), and in layer VI during the first postnatal week (white arrowheads) (F, G). Expression in the cingulate cortex is detected postnatally (open arrowheads F–G) Scale bars, 50 μm (A, B, C), 20 μm (A′-A‴, B′-B‴, C′-C‴), 50 μm (D–E), 200 μm (F), 500 μm (G).
We next defined the temporal course of Fog2 expression in the cortex. FOG2 is first expressed by postmitotic neurons at E12.5, but is not expressed by progenitors (Figure 3D). At E13.5–E15.5, Fog2 is strongly expressed in the cortical plate. By E16.5– E17.5, when CThPN axons extend through the internal capsule to reach the thalamus, Fog2 is strongly expressed in the lower half of the cortical plate (Figure 3E, Figure S2 A– C). During the first postnatal week Fog2 is highly expressed in layer VI, and expression progressively appears in the cingulate cortex (Figure 3 F, G, Figure S2 A–C). After P7, it becomes less restricted to layer VI, and its expression level greatly decreases. Fog2 is not expressed by cortical interneurons or glia (data not shown). Fog2 is also expressed in forebrain regions including amygdala, hippocampus, piriform and entorhinal cortex, hypothalamus, and thalamic reticular nucleus (Figure S2 A–C).
These results demonstrate that Fog2 is a CThPN-specific transcriptional regulator during early neocortical development. Its temporal and subtype-specific expression strongly suggested that Fog2 might function in CThPN postmitotic differentiation.
Loss of Fog2 function specifically disrupts corticothalamic projections in the frontal cortex
To investigate potential functions of Fog2 in CThPN development, we performed loss-of-function studies. Since global Fog2−/− mice die around E13.5 (Tevosian et al., 2000), we generated cortex-specific Fog2−/− mice (Fog2 cKO) to avoid cardiac lethality, by crossing Fog2floxed mice (Manuylov et al., 2007) with Emx1Cre mice. ISH confirmed loss of Fog2 exclusively in the cortex (Figure S3 A–B).
We first studied cortical cytoarchitecture in Nissl stained sections of Fog2 cKO and wild-type (WT) P6 brains. There is no difference in cortical layering, thickness, or appearance of main fiber tracts, such as internal capsule, corpus callosum, or anterior commissure (Figure S3 C–D). There is also no detectable difference in expression of layer VI markers Tbr1, Tle4, and Foxp2 (Figure S3 E–G, I–K). These data identify that lack of FOG2 does not result in gross disruption of cortical organization or layer VI formation, and indicate that Fog2 function is not required for early fate specification of CThPN.
Fog2 is strongly expressed by CThPN at late embryonic and early postnatal stages, when corticothalamic axons are extending through the internal capsule and targeting thalamus. In each cortical area, CThPN are functionally specialized, and project reciprocally and topographically to the appropriate thalamic nuclei that provide thalamic input to that cortical area, establishing distinct feedforward/feedback circuits. We hypothesized that Fog2 might regulate precise CThPN connectivity.
To test this hypothesis, we examined the corticothalamic projections from functionally distinct cortical areas (associative, motor, and primary sensory) to discrete thalamic nuclei in Fog2 cKO mice. We retrogradely labeled CThPN specifically in the frontal cortex (including prefrontal and motor areas) or somatosensory cortex from their corresponding thalamic nuclei: Mediodorsal (MD) for prefrontal cortex, Ventral anterior/Ventral lateral nuclei (VA/VL) for motor cortex, and Ventrobasal complex (VB) for somatosensory cortex (Figure S3 M–R). We systematically quantified labeled CThPN in the dorsal and lateral aspects of frontal and somatosensory cortex at P6 (Figure 4).
Figure 4. Loss of Fog2 function disrupts axonal targeting of CThPN in the frontal cortex.
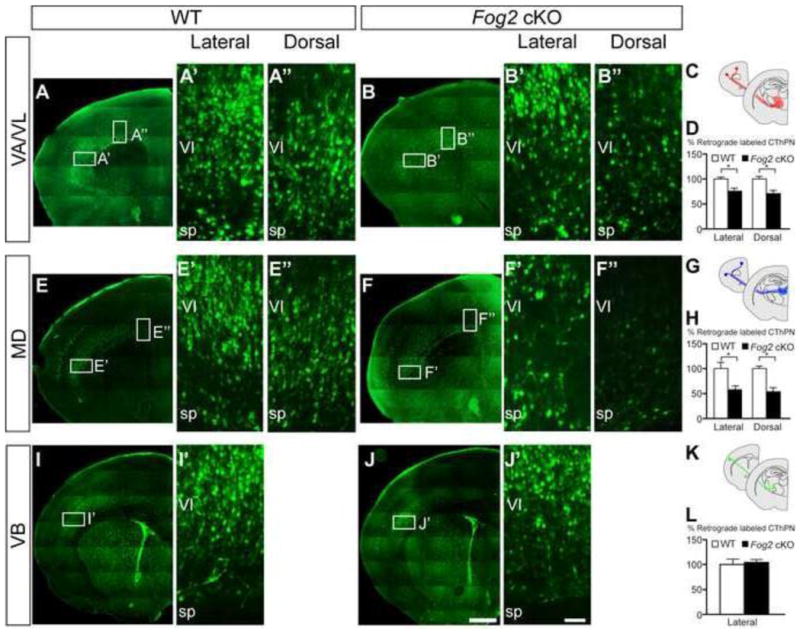
Coronal sections of brains injected with FG in discrete thalamic nuclei: VA/VL (A–B″), MD (E–F″), and VB (I–J′). Boxed areas in (A, B, E, F, I, J) are magnified in (A′–J′). Schematic of the nuclei injected, and the cortical areas where retrograde labeled CThPN were analyzed (C, G, K). Quantification of labeled CThPN in the dorsal and lateral aspects of motor cortex from each nucleus (D, H, L). Asterisk indicates statistical significance (p<0.05, t-test). Scale bar, 500 μm (A–J), 50 μm (A′–J′). Data are represented as mean ± SEM.
We identify a striking reduction in the number of CThPN innervating their targets in Fog2 cKO frontal cortex relative to control littermates (Figure 4 A–H), but no difference in somatosensory cortex (Figure 4 I–L). Interestingly, we find a reduced number of labeled CThPN in frontal cortex from both thalamic targets (MD and VA/VL), and, in both cases, there is a trend toward higher reduction in dorsal than lateral areas (Figure 4 A–D, 32% and 27% reduction in CThPN projecting to VA/VL in dorsal and lateral motor cortex; Figure 4 E–H, 46% and 42% reduction in CThPN projecting to MD in dorsal and lateral frontal cortex). We confirmed that this reduction in labeled CThPN is not due to increased cell death by ICC for cleaved caspase-3 (data not shown). These results indicate that loss of Fog2 function disrupts CThPN connectivity specifically in frontal cortex; within this area, loss of FOG2 might preferentially affect CThPN located dorsally, regardless of their thalamic target, indicating that Fog2 function is subject to precise area-specific regulation.
Fog2 −/− neurons abnormally acquire mixed CThPN/SCPN molecular identity, and extend aberrant axonal projections
Aberrant CThPN axonal targeting in Fog2 cKO mice might be the result of dysregulated identity acquisition. Many core elements of layer VI molecular identity are preserved with loss of Fog2 function, based on normal expression of Tbr1, Tle4, and Foxp2. We hypothesized that imprecise CThPN differentiation might occur by inappropriate expression of controls over development of alternative neuron subtypes, resulting in mixed or imprecise molecular and hodological identity. Fezf2 and Ctip2 are normally expressed at low level by CThPN, and at high level by SCPN (Arlotta et al., 2005; Molyneaux et al., 2005), controlling central aspects of CFuPN specification and connectivity respectively, in a dose-dependent manner. We investigated whether these distinct and critical expression levels are maintained by CThPN in Fog2 cKO cortex.
There is a significant increase in the number of layer VI neurons expressing high level Ctip2 in frontal cortex of Fog2 cKO mice, but not in sensory areas (Figure 5 A, D, J, M). In contrast, there is no difference in Fezf2 expression (Figure S3 H, L), indicating that Fog2 specifically regulates Ctip2, and not the genetically upstream control Fezf2. We performed qPCR to quantitatively investigate Ctip2 expression in frontal cortex. We find ≈1.4 fold upregulation of Ctip2 in Fog2 cKO cortex compared to WT (Figure 5 I). To investigate whether layer VI neurons expressing high level Ctip2 might be presumptive layer V SCPN with abnormal migration, we performed BrdU-birthdating by injecting a single pulse of BrdU at E12.5 or E13.5 (peaks of layer VI-CThPN and layer V-SCPN generation, respectively), and analyzed the distribution of BrdU-positive cells across layers at P6. Since primary motor cortex is an area with strikingly abnormal regulation of Ctip2 and CThPN targeting, we focused our analysis in this area. There is no difference between WT and Fog2 cKO cortex in the number or distribution of BrdU+ neurons at P6 that were labeled at E12.5 or E13.5, indicating that neuronal generation and migration are not altered (Figure 5G, Figure S4). Combined BrdU birthdating at E12.5 with ICC for CTIP2 identifies that 32% of neurons born at E12.5 express high level Ctip2 in layer VI of Fog2 cKO motor cortex, compared to 5% in wild type mice (Figure 5H). The percentage of neurons born at E12.5 that express high level Ctip2 in layer V, normally the earliest generated SCPN, is not significantly different, indicating that loss of Fog2 function specifically affects CThPN (Figure 5H: WT, 40%; Fog2 cKO, 37%; p>0.05). In addition, there are more neurons that co-express Tbr1 and high level Ctip2 in Fog2 cKO motor cortex (Figure 5 J–O), further indicating that motor cortex CThPN undergo aberrant differentiation, and acquire mixed CThPN/SCPN molecular identity.
Figure 5. In the absence of Fog2 function, CThPN are born and migrate normally, but aberrantly up-regulate Ctip2 expression in motor cortex.
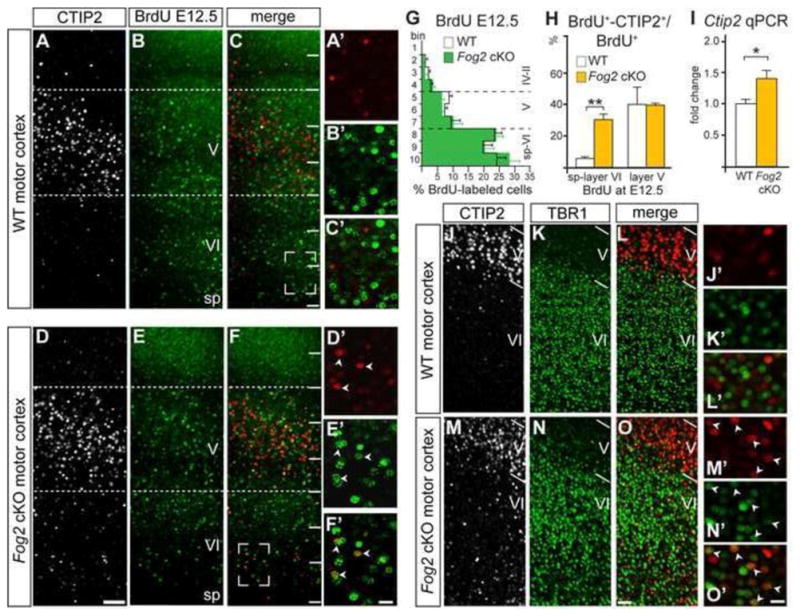
(A–F′) CTIP2 and BrdU ICC on P6 motor cortex. More layer VI neurons BrdU-pulsed at E12.5 express high level Ctip2 in motor cortex of Fog2 cKO mice. (G) Quantification of BrdU pulsed neurons at E12.5 across the thickness of motor cortex shows no differences in the number and distribution of CThPN between WT and Fog2 cKO. (H) Quantification of double labeled BrdU+-CTIP2+ neurons located in layers VI and V expressed as percentage of total BrdU+ pulsed neurons at E12.5. (I) Quantification of Ctip2 expression level in frontal cortex by qPCR analysis. Asterisk (p<0.05), two asterisks (p<0.01). (J–O′) CTIP2 and TBR1 ICC on P6 motor cortex. More neurons in Fog2 cKO motor cortex co- express high level Ctip2 and Tbr1. Scale bars, 100 μm (A–F), 50 μm (J–O), 20 μm (A′–F′, J′–O′). Data are represented as mean ± SEM.
Changes in molecular identity and connectivity observed in motor cortex suggest that CThPN undergo an area-specific change toward SCPN-like identity in the absence of Fog2. To investigate whether CThPN extended axons subcerebrally, we injected FG into the cerebral peduncle at P1, and analyzed the location of retrogradely labeled neurons in motor cortex at P6. Neither the number nor position of SCPN changes in Fog2 cKO cortex, indicating that CThPN do not develop projections to the brainstem (Figure S5). Together, these data indicate that specific CThPN subsets acquire mixed CThPN/SCPN molecular identity, but do not undergo a complete identity change into SCPN.
To further understand the changes in axonal connectivity and molecular identity caused by loss of Fog2 function, we introduced low titer Cre-recombinaseGfp or controlGfp plasmids by in utero electroporation into E11.5 Fog2flx/flx cortex, targeting progenitors that normally generate subplate and CThPN. This approach enables precise analysis of the axonal trajectories and molecular identities of a select, small number of Fog2 KO neurons (CreGfp) in an otherwise normal background. Consistent with the results above, these experiments identify that, while the majority of Fog2 KO neurons target the thalamus, some aberrantly extend their axons into the ventral telencephalon, where they extend along the ventral border of the brain into the hypothalamus up to the midline (Figure 6E–H). Such aberrant projections are never observed in controlGfp neurons (Figure 6 A–D). Importantly, a significant number of CreGfp neurons in layer VI express high level Ctip2 in the most rostral portions of the electroporated area (presumptive motor cortex), while controlGfp electroporated neurons express very low level Ctip2 (Figure 6 B–B‴ and F–F‴). Together, these results strongly support the interpretation that Fog2 is necessary to negatively regulate expression of Ctip2 to the appropriate low dose characteristic of CThPN, and that failure of this regulation results in aberrant CThPN differentiation.
Figure 6. Fog2 −/− neurons upregulate CTIP2 and project aberrantly to the ventral telencephalon.
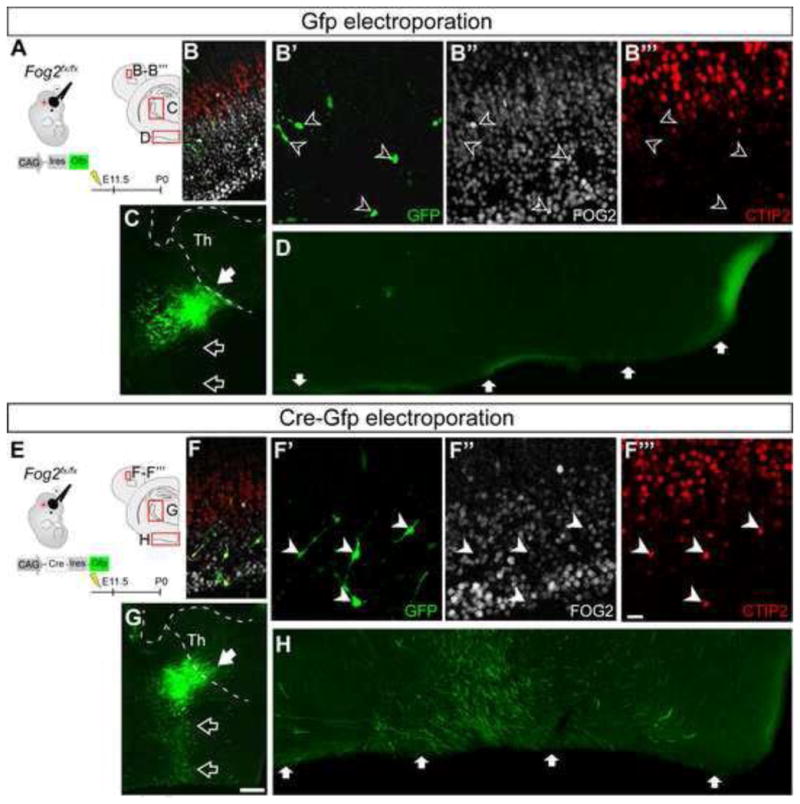
Electroporation of controlGfp or Cre-recombinaseGfp plasmids into Fog2fx/fx embryos at E11.5. (A, E) Schematic of experimental approach, and diagram of the cortical and diencephalic regions shown with immunofluorescence. ICC for GFP, FOG2 and CTIP2 in controlGfp (B′-B‴), and CreGfp electroporated (F-F‴) neurons. In motor cortex, control GFP+ neurons express FOG2, and low level or no CTIP2 (open arrowheads, B-B‴). In contrast, many CreGfp neurons express high level of CTIP2, and do not express FOG2 (white arrowheads, F-F‴). GFP ICC reveals axonal projections from electroporated neurons in the internal capsule en route to thalamus in control mice (C–D), and in the internal capsule, but aberrantly extending into the ventral hypothalamus, in experimental mice (G–H). Scale bars, 20 μm (B-B‴ and F-F‴) and 500 μm (C, G).
Mis-expression of Fog2 Disrupts Differentiation of Subcerebral Projection Neurons
Loss of Fog2 causes aberrant axonal targeting and abnormal regulation of Ctip2 expression by motor CThPN, modifying their differentiation to acquire some cardinal features of SCPN. To investigate whether mis-expression of Fog2 in SCPN conversely disrupts SCPN development, including expression of Ctip2 and axonal projections, we performed gain-of-function experiments.
We co-electroporated tdTomato and Cre-recombinase plasmids at high and low titers, respectively, into E13.5 embryos encoding a CAG-floxed-Stop-Fog2-IRES-Gfp cassette in the Rosa26 locus (CTV-Fog2 mice). This creates a mosaic of electroporated cells in which sparse Fog2 mis-expressing neurons (CRE+, FOG2+, GFP+, and tdTomato+) are scattered among control (tdTomato+) neurons (Figure 7A–F, Figure S6 A–D). Since CThPN and SCPN are generated consecutively with overlap in their production, electroporations at E13.5 that predominantly target SCPN also target a subpopulation of the last generated CThPN, whose axonal projections must be considered during analysis of Fog2 effects on axonal connectivity of SCPN. To rigorously correct for this late-born CThPN contribution to the projections analyzed, we quantified CThPN and corticothalamic axons across the electroporation area, and calculated the ratio of CTh axons/Layer VI neurons, which should be approximately equal to one in controls (Figure 7A–C). If Fog2 mis-expression were to redirect SCPN axons toward the thalamus, more CTh axons would exist than genuine CThPN; therefore the ratio GFP+ CTh axons/Layer VI GFP+ neurons should be higher than in controls. These experiments find that the ratio of GFP+ CTh axons/Layer VI GFP+ neurons is not significantly different than in the controls, indicating that Fog2 mis-expression in SCPN does not redirect axons toward thalamus (Figure 7B). However, Fog2 mis-expression consistently causes reduced expression of Ctip2 in layer V SCPN (Figure 7C–K). We quantified the percentage of tdTomato+ and GFP+ neurons expressing Ctip2 within layer V (Figure 7K). There is a significant decrease in the proportion of Fog2 mis-expressing neurons that express Ctip2 relative to control, indicating that Fog2 represses expression of Ctip2 by SCPN (64% reduction in the number of CTIP2/GFP+ neurons compared to control CTIP2/tdTomato+ neurons, p<0.05). Further, significantly fewer axons of Fog2 expressing neurons compared to controls reach caudal internal capsule and cerebral peduncle (Figure 7 L–O). These abnormal projections are reminiscent of the strikingly reduced and dysmorphic axonal projections to subcerebral targets observed in Ctip2 KO mice (Arlotta et al., 2005). To further investigate whether Fog2 functions in part by regulating Ctip2 expression, we performed mis-expression of Fog2 in CPN. There is no change in CPN axonal projections (data not shown). Together, these results indicate that Fog2 is critical in regulating Ctip2 expression to control proper CThPN development, rather than simply inducing corticothalamic projections by other neuron subtypes.
Figure 7. Fog2 mis-expression in SCPN reduces expression of Ctip2 and subcerebral axon projections, but does not induce projection to thalamus.
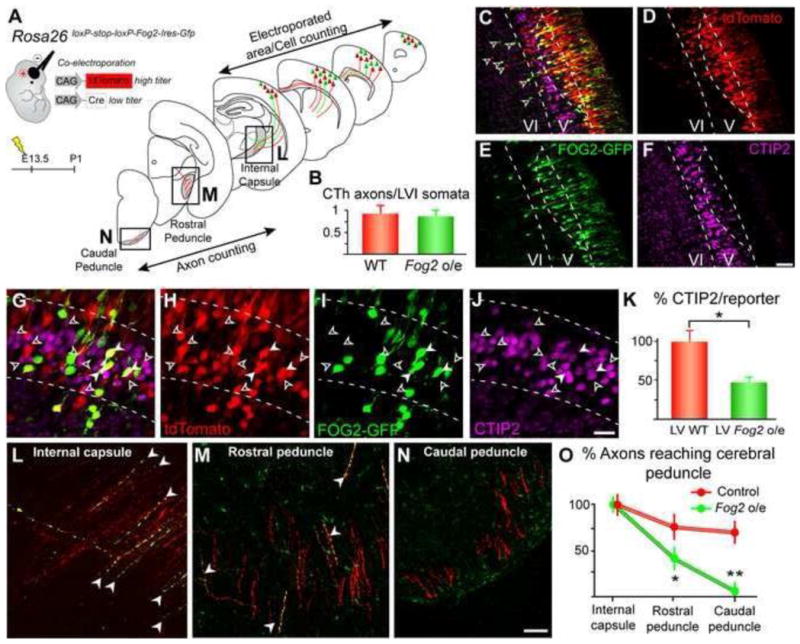
(A) Schematic of experimental approach, and diagrams of the brain regions where electroporated neuron somata and axons were quantified. Electroporations always targeted motor cortex, but also spread into somatosensory cortex. (B) Quantification of corticothalamic axonal projections upon Fog2 mis-expression. The number of CTh axons is not significantly higher than the number layer VI-CThPN after electroporation at E13.5 (open arrowheads in C), in control or experimental conditions. (C–K) Fog2 mis- expression in layer V (between dashed lines) reduces expression of Ctip2 by SCPN. (G– K) The number of FOG2-GFP+ neurons expressing Ctip2 (white arrowheads) is significantly reduced compared to the number of tdTomato-only neurons expressing Ctip2 (open arrowheads). (K) For quantification, values are normalized to the ratio CTIP2/reporter in control tdTomato-only neurons (control CTIP2/tdTomato-only 100%, experimental CTIP2/GFP+ 46%). Asterisks indicate statistical significance (p<0.05). (L–O) Axons of SCPN mis-expressing Fog2 progress significantly less far in the cerebral peduncle than axons of control SCPN. Quantification of subcerebral projecting axons was performed at three levels: internal capsule (L), rostral cerebral peduncle (M), and caudal cerebral peduncle (N). Numbers of axons at each level are normalized to the initial number of axons in the internal capsule (O). Asterisk (p<0.05), two asterisks (p<0.01). Scale bars, 50 μm (C–F) and 20 μm (G–N). Data are represented as mean ± SEM.
FOG2 regulates Ctip2 expression level via context-specific transcriptional complexes
The loss-of-function and gain-of-function studies above indicate that Fog2 regulates Ctip2 expression. Interestingly, while all CThPN express Fog2 across the neocortex, only a specific subset in frontal motor cortex upregulate Ctip2 expression and develop abnormal projections in Fog2 cKO mice. Therefore, we investigated mechanisms by which Fog2 regulates Ctip2 expression, and whether this regulation might vary in an area-specific manner.
Since FOG2 lacks a DNA binding domain, its transcriptional activity is highly dependent on interactions with other controls, which might be differentially expressed across areas and neuron subtypes. FOG2 interacts with members of the Coup-tf and Gata families of transcription factors, forming transcriptional complexes with critical functions in the development of other organs (Tevosian et al., 2000; Ackerman et al., 2007). COUPTF1, COUPTF2, GATA2, and GATA4 are expressed in the developing cortex (Tripodi et al., 2004; Minegishi et al., 1999; Agnihotri et al., 2009), and multiple repeats of their consensus binding site sequences map to a highly conserved region upstream of Ctip2 (Figure 8 G), suggesting that these factors might interact with FOG2 in transcriptional complexes to regulate Ctip2 expression.
Figure 8. FOG2 interacts with COUPTF1, GATA2, and GATA4 to regulate expression of Ctip2.
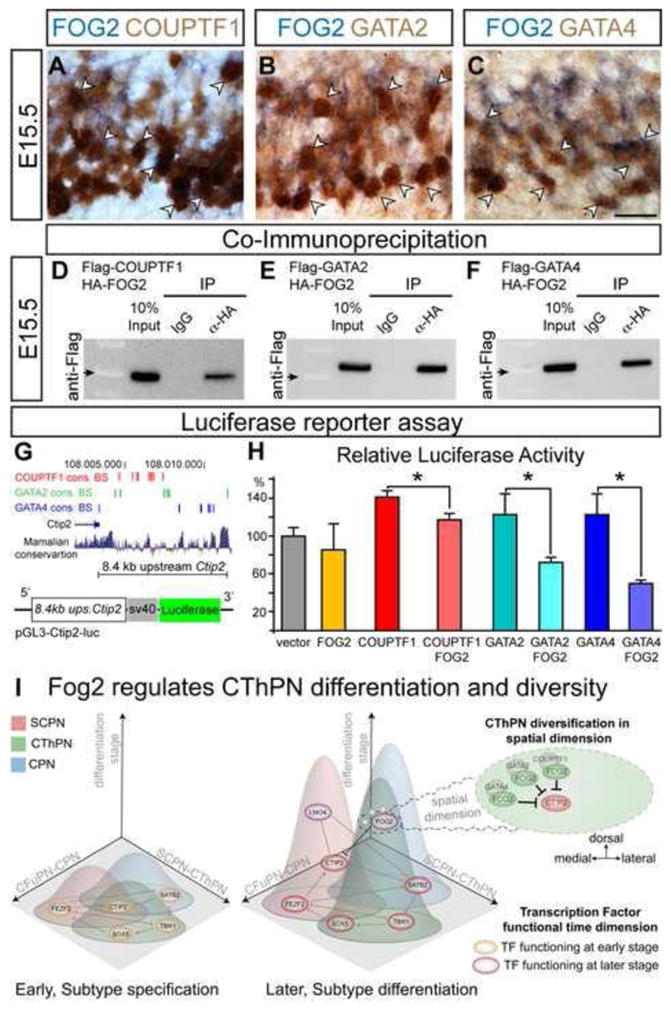
(A–C) Co-expression of FOG2 and COUPTF1, GATA2, and GATA4 in the developing motor cortex identify by ICC for COUPTF1, GATA2, and GATA4 combined with ISH for FOG2 at E15.5. Examples of double positive cells are shown (arrowheads). (D–F) Co-IP of FOG2 with COUPTF1 (D), GATA2 (E), and GATA4 (F). Immunoblotts with Anti-Flag antibody detect Flag-COUPTF1, Flag-GATA2, and Flag-GATA4 proteins pulled-down by anti-HA antibody, but not by non-specific IgG. Arrow marks 50KDa on the ladder to estimate protein size. (G) Putative regulatory region upstream of Ctip2 used for luciferase reporter assays contains COUPTF1, GATA2, and GATA4 consensus binding sites (red, green, blue, respectively) and is highly conserved across mammals. (H) Percentage of luciferase activity relative to the baseline (pGL3-Ctip2-Luciferase alone) in the presence of FOG2, COUTF1, COUPTF1 + FOG2, GATA2, GATA2 + FOG2, GATA4, GATA4 + FOG2. Asterisks indicate significance (t-test; p<0.05). Data are represented as mean ± SEM. (I) Schematic depicting CThPN subtype development, throughout distinct differentiation stages, from subtype specification to functional specialization. CPN, SCPN, and CThPN are schematized within an n-dimensional ‘subtype identity space’ in which individual subtype identities are defined by a range of variables (e.g. neuron morphology, axonal projections, circuit connectivity, three-dimensional spatial location, gene expression, the time of function of molecular controls, and other characteristics). This n-dimensional subtype identity is controlled by distinct transcription factors and other controls acting at progressive differentiation stages (e.g. early subtype specification, orange ovals; later subtype differentiation, purple ovals; some act both early and late, dual color ovals). Controls regulating identity development form key nodes of a transcriptional network, which is beginning to be elucidated. Fog2 is depicted in relationship to other key controls, regulating CThPN differentiation and diversity. Controls acting at the emergence of subtype identity can function in multiple, progressively more complex dimensions (e.g. wiggly lines depict extension into a representation of the CThPN spatial dimension, in which Fog2 functions differentially, repressing Ctip2 expression in dorso-medial CThPN subsets in combination with intersecting factors that might not be specific only to the CThPN subtype context). Basic elements of the left and middle sections of Panel I are modified with permission from Greig et al., 2013.
We first investigated expression of COUPTF1, COUPTF2, GATA2, and GATA4 in developing CThPN by ICC at E12.5 and E15.5 in the preplate and cortical plate of presumptive motor cortex, where loss-of-function studies reveal increased Ctip2 expression. FOG2, COUPTF1, and, GATA2 are expressed across the preplate at E12.5 (Figure S7 A, B, D), and are expressed in the deep cortical plate at E15.5, consistent with the location of developing CThPN (Figure S7 E, F, H). COUPTF2 is expressed by VZ/SVZ progenitors and meningeal cells, but not by dorsal preplate neurons at E12.5, nor by dorsal cortical plate neurons at E15.5 (Figure S7 C, G). GATA4 is not expressed in the cortex at E12.5 (data not shown), though at E15.5 it is highly expressed in the deep medio-dorsal cortical plate, but not laterally (Figure S8 I). GATA4 area-specific expression suggests that it might uniquely interact with FOG2 in the developing frontal motor cortex. We investigated and confirmed co-expression of these factors in the dorsal cortical plate, combining Fog2 ISH with ICC for COUPTF1, GATA2, and GATA4 (Figure 8 A–C).
We next investigated whether FOG2 interacts with COUPTF1, GATA2, and GATA4 by protein co-immunoprecipitation (Co-IP). We generated Flag-Couptf1, Flag- Gata2, Flag-Gata4, and HA-Fog2 expression constructs, and co-transfected HA-Fog2 with each of the three potentially interacting factors in dissociated E15.5 cortical neurons. We performed IP with anti-HA antibody, and blotted with anti-Flag to detect Flag- COUPTF1, Flag-GATA2, or Flag-GATA4. These results identify that COUPTF1, GATA2, and GATA4 are able to form protein complexes with FOG2 in cortical neurons (Figure 8 D–F). GATA2 and GATA4 are able to pull down FOG2 more efficiently than COUPTF1, suggesting that interactions between FOG2 and these GATA factors might be stronger than between FOG2 and COUPTF1.
We then investigated the ability of FOG2 alone, or in combination with COUPTF1, GATA2, or GATA4, to regulate expression from a putative regulatory region spanning 8.4 kb upstream of Ctip2, using luciferase reporter assays. This region contains multiple repeats of COUPTF1, GATA2, and GATA4 consensus binding sites (TRANSFAC dabase, Biobase) (Figure 8 G). We cloned this Citp2 upstream region into a luciferase reporter construct (pGL3-Ctip2-luc, Figure 8 G), and transfected E15.5 cortical neurons with pGL3-Ctip2-luc alone or together with Fog2, Couptf1, Gata2, or Gata4 constructs separately, or pGL3-Ctip2-luc combined with both Fog2 and each individual potentially interacting factor. While neither FOG2, GATA2, or GATA4 alone have a significant effect in regulating expression from the Ctip2 upstream region, FOG2 in combination with GATA2, or GATA4 strongly reduces luciferase activity (49.6% and 59.2% reduction of the activity yielded by GATA2 and GATA4 alone, respectively; Figure 8 H). Strikingly, while COUPTF1 alone increased luciferase activity relative to baseline, FOG2 combined with COUPTF1 moderately reduces activity relative to COUPTF1 alone (23.2% reduction, Figure 8 H). These results identify that FOG2 acts in combination with other controls to regulate Ctip2 expression level. FOG2 forms distinct transcriptional complexes with different efficiencies in regulating Ctip2 expression. While transcriptional complexes containing FOG2-GATA2 or FOG2-COUPTF1 might function across multiple areas, complexes containing FOG2-GATA4 more likely uniquely function in the developing frontal dorsal motor cortex, and likely contribute to the area-specificity of the phenotype observed in the Fog2 cKO cortex.
Together, these studies identify that Fog2 regulates the precise development of corticothalamic subtype identity, in large part by regulating the dosage of CTIP2, sharpening the distinction of CThPN from other corticofugal subtypes, and ensuring appropriate area-specific CThPN differentiation without interference from the SCPN developmental program. FOG2 acts combinatorially and intersectionally with multiple transcriptional regulators, in area-specific combinations, likely producing distinct functional complexes that increase CThPN diversity (Figure 8 I).
Discussion
Here, we report a series of experiments focusing on molecular controls over development of corticothalamic projection neurons to address the broader question of molecular mechanisms that generate neuronal diversity in the neocortex. We identified a set of genes highly enriched in CThPN from their specification through their axonal targeting and refinement; these genes have largely uncharacterized functions in the nervous system, and their expression progressively defines CThPN. Our first functional investigation of whether these identified genes include controls regulating CThPN development identifies the first known function of Fog2 in the nervous system, controlling precision of differentiation and diversity of CThPN. Fog2 is necessary for proper acquisition of projection and molecular identity of motor CThPN, at least in substantial part by regulating the dosage of CTIP2 via distinct interaction with multiple controls. These interactions form multiple specific transcriptional complexes, which might function in an area-dependent, stage-dependent, and/or other context-dependent manner. Such intersecting mechanisms add a new layer of complexity and precision to the CThPN developmental program, not yet taken into account in current models of neuron subtype differentiation. This added complexity and precision likely contributes very importantly to areally and functionally diversify the broad class of CThPN.
Functions of novel CThPN-genes in CThPN development and diversity
Cortical projection neurons progressively acquire subtype and area identity by execution of precisely regulated genetic programs. Genes common to all projection neurons, and subtype-specific genes, sequentially contribute to these programs, interacting in combinatorial fashions to instruct progenitors to differentiate into distinct functional neuron types. Our work here aimed to identify molecular controls over development of CThPN subtype, including specification, differentiation, and acquisition of precise area identity.
We identified a novel set of genes differentially expressed both by the broad class of CThPN, and by distinct CThPN subsets –either expressed in gradients, or restricted to discrete areas or sub-lamina (Figure 1, Figure 2, and Figure S1). These novel CThPN- expressed genes regulate important developmental processes in other cell types, but little to nothing is known about their functions in neuron development, and even less about their likely combinatorial interactions in the context of CThPN development. However, some functional relationships have been described that are potentially highly relevant regarding CThPN differentiation and diversification. For example, a number of the CThPN-genes identified (e.g. Gas7, Klf9, Itsn1, Phr1, Ror2, Shb) have been reported to regulate neurite outgrowth and/or pruning in vitro, and some in vivo. Interestingly, Itsn1 KO mice exhibit axonal defects in the internal capsule and other fiber bundles (Sengar et al., 2013), and in Phr1 KO mice, corticofugal axons are unable to enter the internal capsule (Bloom et al., 2007), strongly suggesting important functions of these genes in CThPN axonal navigation and connectivity.
Importantly for CThPN diversification, we identified Ror2 as a CThPN-specific gene expressed in a latero-medial gradient. The Ror2 promoter is regulated by Foxp2 (Konopka et al., 2009), which is known to be expressed by CThPN and other neuron types. Both Ror2 and Foxp2 have been independently reported to regulate neurite outgrowth and synaptic formation (Paganoni et al., 2005; Reimers-Kipping et al., 2011). It is likely that the combinatorial interaction between a non-CThPN-specific control (e.g. Foxp2) and a CThPN-specific control (e.g. Ror2) might contribute collaboratively to regulate neurite development in a subset of the broad CThPN population, perhaps in a graded manner. Similarly, we identified Cask as a CThPN-specific gene particularly enriched in CThPN located in deeper layer VI. CASK regulates synaptic function and neuronal development, binding to critical transcriptional controls such as TBR1 and CTIP1 (Hsueh 2006; Kuo et al., 2010), both expressed by CThPN and other subtypes (McKenna et al., 2011; Greig et al., 2016). Interestingly, Cask KO mice exhibit abnormalities in synaptic transmission, and increased neuronal death in the thalamus (Atasoy et al., 2007), strongly suggesting important function of Cask, and Cask-related controls, regulating development of synaptic function in specific CThPN subsets.
Of particular interest related to Fog2 function in CThPN are Smarcd3 and Zmiz1, transcriptional regulators identified here as enriched in CThPN subsets. Smarcd3 and Zmiz1 regulate Gata4 and Gata2 functions during cardiomyogenesis and hematopoiesis, respectively (Christoforou et al., 2013; Beliakoff et al., 2008). Given the importance of Gata factors as intersecting controls of Fog2 function, it is likely that Smarcd3 and Zmi1 contribute to refine Fog2 function in CThPN differentiation and diversification through combinatorial interactions with Gata factors. Much work will be needed to more fully understand functions and relationships of molecular controls over CThPN development, diversity, and specialization of circuit function. Here, we identify novel CThPN-specific genes previously not known to be related to CThPN differentiation, opening new avenues for investigating likely multi-dimensional functions and interactions of these controls in neuron subtype development.
CThPN are functionally diverse, but consist of two broad laminar and functional populations. 1) Layer VI-CThPN, which project to all thalamic nuclei, are the most abundant. 2) Layer V-CThPN, which project to specific thalamic nuclei and subcerebral targets simultaneously, account for a very small proportion of all CThPN (Guillery and Sherman 2002). Further diversity of the predominant layer VI-CThPN population is observed in distinct cortical areas, where CThPN are specialized for processing information of distinct modalities (e.g. motor, visual, auditory). Even within a single area, functional differences have been reported within layer VI-CThPN (Briggs et al., 2010). We focused on the predominant layer VI-CThPN population, rather than potentially confounding analysis by inclusion of layer V-CThPN with their dual CThPN/SCPN projecting features. This focused strategy enabled identification of some key elements of the areal and intra-laminar diversity of the main CThPN population. We identified genes differentially expressed by distinct CThPN subsets –either expressed in gradients, or restricted to discrete areas or sub-lamina (Figure 2, Figure S1). Combinatorial interactions of area-specific, sub-lamina-specific, and subtype-specific genes likely refine CThPN identity, generating additional levels of diversity of corticothalamic circuitry. Future studies to identify controls underlying differentiation of layer-specific CThPN populations will be of great interest. This work focuses on the main CThPN population, as a first step toward investigation of overall CThPN diversity, and lays the foundation for future investigation of CThPN diversity across areas, layers, and lamina.
Fog2 function in CThPN differentiation
Fog2 functions as a transcriptional co-repressor in several cell types (Tevosian et al., 2002; Manuylov et al., 2007). Our studies identify a similar co-repressor function in CThPN, regulating the expression level of Ctip2. Our loss-of-function experiments indicate that abnormally high-level Ctip2 expression in CThPN likely induces a “hybrid” identity, with both features of the SCPN developmental program and residual CThPN features. The SCPN features arise in an inappropriate cellular/molecular context (CThPN molecular markers are still expressed), leading to abnormal differentiation and failed projection into the cerebral peduncle.
Expression of the correct level of key molecular controls is critical for appropriate subtype identity acquisition, so regulation of the expression level of these controls is likely both area-specific and neuron subtype-specific; dysregulation of these levels would be expected to result in distinct alterations that depend on the specific cellular context. Cell context-dependency of phenotypes caused by dysregulation of expression levels of Ctip2 and other key molecular controls has been observed and reported repeatedly, supporting this interpretation (Molyneaux et al., 2005; Chen et al., 2008; Kwan et al., 2008; Lai et al., 2008; Han et al., 2011; McKenna et al., 2011). Interestingly, reduced Fog2 expression and concomitant increased Ctip2 expression in layer VI has been reported in Sox5 KO mice (Kwan et al., 2008, Lai et al., 2008), and recently in Ctip1 KO mice (Woodworth et al., 2016; in particular, Fig 3). Importantly, in Ctip1 KO mice, corticothalamic innervation is significantly reduced (Woodworth et al., 2016; in particular, Fig 4 G–O); and in Sox5 KO mice, axon fascicles aberrantly extend into the ventral telencephalon and hypothalamus (Kwan et al., 2008 Fig 5; Lai et al., 2008 Fig 3), strikingly resembling the misrouted axons reported here in the ventral telencephalon in Fog2 cKO mice. Given the critical role of Ctip2 expression levels in corticofugal differentiation and axonal development and connectivity, these independent observations strongly support the interpretation that Fog2 regulation of Ctip2 expression is crucial for appropriate CThPN development. Together, these results highlight the importance of regulating dosage of key controls for precision of differentiation, and the critical role of Fog2 in refining CThPN identity in functionally specialized cortical areas.
Area-specific regulation of Fog2 function
While Fog2 is expressed by CThPN across cortical areas, the abnormalities observed in Fog2 cKO mice are restricted to frontal cortex. A variety of mechanisms might contribute to control of this area-specific Fog2 function, including area-specific control of protein-protein interactions and post-translational modifications.
FOG2, like other transcriptional co-regulators, functions in combination with other transcriptional controls. We identify that FOG2 is able to interact with GATA2, GATA4, and COUPTF1 in developing cortical neurons; when combined with these controls, FOG2 regulates Ctip2 expression in vitro. Each of these interaction pairs is known to function critically in other systems. For example, FOG2-GATA4 complex is required for cardiac, gonadal, and lung development (Tevosian et al., 2000; 2002; Ackerman et al., 2007). Importantly, we report that FOG2 and GATA4 co-express in developing CThPN in motor cortex, and that FOG2-GATA4 strongly downregulate expression of Ctip2 in vitro. Thus, area-specific expression of GATA4 very likely contributes to control FOG2 function in an area-specific manner.
GATA2, best studied in hematopoiesis development, is expressed across the cortical plate, and might potentially interact with FOG2 across areas. FOG2-GATA2 strongly regulates expression of Ctip2 in vitro. Interestingly, GATA factors are able to simultaneously bind Fog and Lmo (LIM-domain only) co-regulators in multiprotein complexes to regulate transcription (Wilkinson-White et al., 2011). Lmo regulators are also expressed in the cortex. Importantly, Lmo4 is expressed in an area- and subtype- specific fashion, and is a critical control over projection neuron diversity in the rostral motor cortex (Cederquist et al., 2013). Additionally, recent studies identify a novel function of Lmo4 regulating Ctip2 expression in a specific subset of Satb2 expressing neurons (Harb et al., 2016). This suggests that Lmo4 might contribute to regulate FOG2- GATA interactions, by a similar context-specific regulatory mechanism.
Our studies identify that FOG2-COUPTF1 modestly but significantly regulates Ctip2 expression in vitro. Also, our results indicate that COUPTF1 is able to pull down FOG2 less efficiently, compared to GATA factors, suggesting either weaker interaction or the presence of competing factors rendering COUPTF1 less available to interact with FOG2. Interestingly, CTIP1 and CTIP2 (Coup-Tf Interacting Protein 1 and 2), and FOG2 have common structural domains, such as the amino terminal repressor domain, and the CCHH zinc fingers able to interact with COUP-TFs (Huggins et al., 2001; Lin, et al., 2004). Ctip1 is highly expressed by CThPN in sensory cortices, but only weakly expressed by CThPN in motor cortex (Greig et al., 2016). It is possible that CTIP1 regulates FOG2 interaction with COUP-TF1 in an area-specific manner, and/or compensates for Fog2 function in sensory areas.
Post-translational modifications might also contribute to control of Fog2 area-specific function. Sumoylation is a highly dynamic post-translational modification that regulates the activity state of transcription factors in distinct cellular contexts. Importantly, sumoylation regulates FOG2 repressive activity in cardiomyocytes (Perdomo et al., 2012), and might similarly regulate the repressive function of FOG2 in neurons.
FOG2 and human intellectual disability
Abnormal assembly of cortical circuits is hypothesized to underlie many neurodevelopmental disorders such as cognitive disability, autism spectrum disorders, epilepsy, schizophrenia, and ADHD (Kwan 2013, Mitchell 2011). Here, we report that, in the absence of Fog2 function, CThPN circuitry is abnormal in the prefrontal and motor cortex. Fog2 is expressed in human developing neocortex (BrainSpan: Atlas of the Developing Human Brain www.brainspan.org). In humans, Fog2 mutations have been primarily studied in the context of congenital heart diseases, such us Tetralogy of Fallot (TOF), and congenital diaphragmatic hernia (CDH). Interestingly, in TOF and CDH patients, behavioral and/or neurological phenotypes are commonly reported (Piran et al., 2011). Hand sterotypies, delayed or no speech acquisition, seizures, and mild to severe intellectual disability have been reported in patients with deletions involving Fog2 (Thierry et al., 2013; Wat et al., 2011). Whether Fog2 variation might contribute to some human intellectual disabilities will be of substantial interest to investigate further.
Experimental Procedures
Animals
All mouse studies were approved by the Harvard University IACUC, and were performed in accordance with institutional and federal guidelines. Further details are listed in Supplemental Experimental Procedures.
Affymetrix Microarrays
CThPN were retrogradely labeled and FACS-purified at E18.5, P3, and P6. RNA extraction, hybridization into Affymetrix GeneChips, and microarray data analysis was performed as described in Arlotta et al., 2005. Microarray data have been deposited in the GEO database (Accession GSE61711). Further details are listed in Supplemental Experimental Procedures.
Immunocytochemistry and in situ hybridization
Brains were immunostained using standard methods (Molyneaux, et al., 2005). Antibodies are listed in Supplemental Experimental Procedures. ISH was performed as described in Arlotta et al., 2005. Primers for riboprobes are listed in Supplemental Table S2.
Retrograde Labeling
Projection neurons were retrograde labeled by ultrasound guided injection of FluoroGold at P1 to P3. Further details are listed in Supplemental Experimental Procedures.
BrdU birthdating
BrdU birthdating and quantification was performed as described in (Molyneaux et al., 2005). Further details are listed in Supplemental Experimental Procedures.
In utero electroporation
Electroporation were performed as described in Molyneaux et al., 2005. Further details about procedure and plasmids are provided in Supplemental Experimental Procedures
Co-Immunoprecipitation
Co-IP was performed using Protein-A/G beads from E15.5 cortical neuron protein extracts following standard methods (Huggins et al., 2001). Further details about neuronal transfection, plasmids, and antibodies are provided in Supplemental Experimental Procedures.
Luciferase assays
Luciferase reporter assay was performed and analyzed as described in McKenna et al., 2011. Further details are provided in Supplemental Experimental Procedures.
Supplementary Material
Figure S1. (related to Figure 1) Genes identified by microarray analysis are expressed in the cortical plate and layer VI.
In situ hybridization or immunocytochemistry showing expression in layer VI for all genes selected from Figure 1 (except genes shown in Figure 2). For each gene, the temporal course of gene expression from microarray analysis by CThPN (green), CSMN (red), and CPN (blue) are shown in the same panel. Expression is shown at E16 or postnatal stages (P0, P1, or P3). For genes expressed by CThPN subpopulations (Zmiz1, Cask, Dmrt2, Phr1, Zmynd11), arrowheads indicate restricted expression. Immuncytochemistry for CASK detects CThPN in lower layer VI, and CThPN axons in the internal capsule (white arrowheads). Scale bars, 250 μm (E16), 150 μm (P0) and 500 μm (P1, P3).
Figure S2. (related to Figure 3) Fog2 is expressed by CThPN across cortical areas.
(A–C) Fog2 is strongly expressed in layer VI by CThPN, and by other neuron subtypes in other forebrain regions. (D–I) In the cortex, FOG2 is excluded from high level CTIP2- expressing neurons (SCPN) (D–F), and SATB2-expressing neurons (CPN, arrows) (G–I). (J–N) FOG2 expression is CThPN specific, compared to TBR1, also expressed in layer VI CPN (white arrowheads in L–N). Scale bars, 500 μm (A–C), 100 μm (D–F, J), 20 μm (G–I) and 10 μm (L–N).
Figure S3. (related to Figure 4) Cortical morphology and expression of widely used layer VI markers are not affected by the absence of Fog2 function in cortex. (A–B) Fog2 cKO mice show absence of Fog2 mRNA specifically in the neocortex (layer VI, black arrowheads), but show normal expression in other forebrain regions. (C–D) Nissl staining of cortex in wild-type (C) and Fog2 cKO (D) mice at P7 reveals no differences in laminar patterning. (E–L) IHC for TBR1 (E, I), TLE4 (F, J), FOXP2 (G, K), and ISH for Fezf2 (H, L) show no difference in expression of these markers between WT and Fog2 cKO cortices. (M–R) FluoroGold injections in thalamic nuclei in WT and Fog2 cKO brains: (M, P) Mediodorsal nucleus (MD); (N, Q) Ventral anterior/Ventro lateral nucleus (VA/VL); and (O, R) Ventrobasal complex (VB). Scale bar, 50 μm (E–K) and 500 μm (M–R).
Figure S4. (related to Figure 5) In the absence of Fog2 function, SCPN are born and migrate normally. (A–C) Quantification of E13.5 BrdU birthdated neurons at P6 across the thickness of motor cortex. There is no difference between WT and Fog2 cKO cortex in the number or distribution of BrdU+ neurons at P6 that were labeled at E13.5, indicating that SCPN generation and migration are not altered in Fog2 cKO mice. Scale bar, 100 μm (A–B). Data are represented as mean ± SEM.
Figure S5. (related to Figure 6) Axonal projections to the cerebral peduncle are normal in the absence of Fog2 function. (A–E) SCPN are correctly located in layer V, and not in layer VI, in motor cortex of Fog2 cKO mice. (A) Schematic of experimental approach. (B, C) Low magnification images of the area analyzed. (D–E) High magnification of area boxed in B–C. Scale bar, 60 μm (D–E).
Figure S6. (related to Figure 7) Fog2 is mis-expressed exclusively in Cre-electroporated neurons in CTV-Fog2 embryos (CAG-floxed-Stop-Fog2-IRES-Gfp). (A–D′) Cre and tdTomato co-electroporation in CTV-Fog2 embryos at E13.5 results in Fog2 mis- expression in layer V and upper-layers, only in Cre-recombined neurons (GFP+, FOG2+ detected by immunolabeling; white arrowheads in A′–D′). FOG2 is not expressed in tdTomato-only electroporated neurons (open arrowheads, A′–D′). (A′–D′) High magnification of area boxed in A–D. Scale bars, 50 μm (A–D) and 25 μm (A′–D′).
Figure S7. (related to Figure 8) COUPTF1, COUPTF2, GATA2, and GATA4 expression in the developing cortex. Immunocytochemistry for FOG2, COUPTF1, and GATA2 shows expression of these proteins in the preplate of developing cortex at E12.5 (A, B, D), and in the cortical plate at E15.5 (E, F, H). Higher magnification images of the boxed areas corresponding to developing motor cortex are shown in (A′– H′). Immuncytochemistry for COUPTF2 shows expression primarily by progenitors and meningeal cells, but not in the preplate at E12.5 (C-C′), nor in the dorsal cortical plate at E15.5 (G-G′). Immuncytochemistry for GATA4 shows expression at E15.5 in a medio-lateral gradient, with highest expression in the medial and dorsal cortical plate, but little or no expression the lateral cortical plate (I-I′). No expression was detected at earlier time points (not shown). Scale bars, 20 μm (A′–I′).
Supplemental Table 1. (related to Figure 1) Summary of Gene Ontology terms for biological functions over-represented among CThPN-genes.
Supplemental Table 2. (related to Figure 1 and Figure 2) cDNA clones for riboprobes.
Acknowledgments
We thank B. J. Molyneaux for invaluable help setting up retrograde labeling, FACS, microarray experiments, and for insightful comments on earlier versions of this manuscript; P. Arlotta and D. Jabaudon for important early assistance with microarray analysis; S. Tevosian and N. Manuylov for generously sharing Fog2floxed mice before publication; K. Quinn, A. Palmer, T. Yamamoto, K. Billmers, and E.L. Gornstein for technical assistance; D. Dombkowski for assistance with FACS; L. Goodrich for generously sharing CTV-Fog2 mice; C. Lois for plasmids; H. Padmanabhan, V. Sahni, and J.L. MacDonald for enlightening scientific discussions; and other members of the Macklis lab for helpful suggestions.
This work was supported by grants from the NIH (R01NS45523, with additional infrastructure support by R37NS41590, R01NS49553, R01NS75672), and the Harvard Stem Cell Institute. M.J.G. was partially supported by fellowships from CajaMadrid Foundation and the Spanish Ministry of Education. J.G.E. was partially supported by fellowships from the Heart and Stroke Foundation of Canada and the Paralyzed Veterans of America/Travis Roy Foundation.
Footnotes
Supplemental Information includes Supplemental Experimental Procedures, seven figures, and two tables.
Author Contributions
M.J.G., J.G.E., and J.D.M. conceived the project and designed experiments; M.J.G. and J.G.E. performed the experiments; M.J.G., and J.D.M. analyzed and interpreted the data; and M.J.G. made the figures. M.J.G. and J.D.M. synthesized and integrated the findings and wrote and revised the paper. All authors read and approved the final manuscript.
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- Ackerman KG, Wang J, Luo L, Fujiwara Y, Orkin SH, Beier DR. Gata4 is necessary for normal pulmonary lobar development. Am J Respir Cell Mol Biol. 2007;36:391–397. doi: 10.1165/rcmb.2006-0211RC. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Agnihotri S, Wolf A, Picard D, Hawkins C, Guha A. GATA4 is a regulator of astrocyte cell proliferation and apoptosis in the human and murine central nervous system. Oncogene. 2009;28:3033–46. doi: 10.1038/onc.2009.159. [DOI] [PubMed] [Google Scholar]
- Arlotta P, Molyneaux BJ, Chen J, Inoue J, Kominami R, Macklis JD. Neuronal subtype-specific genes that control corticospinal motor neuron development in vivo. Neuron. 2005;45:207–221. doi: 10.1016/j.neuron.2004.12.036. [DOI] [PubMed] [Google Scholar]
- Atasoy D, Schoch S, Ho A, Nadasy KA, Liu X, Zhang W, Mukherjee K, Nosyreva ED, Fernandez-Chacon R, Missler M, Kavalali ET, Südhof TC. Deletion of CASK in mice is lethal and impairs synaptic function. Proc Natl Acad Sci U S A. 2007;104:2525–2530. doi: 10.1073/pnas.0611003104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bedogni F, Hodge RD, Elsen GE, Nelson BR, Daza RA, Beyer RP, Bammler TK, Rubenstein JL, Hevner RF. Tbr1 regulates regional and laminar identity of postmitotic neurons in developing neocortex. Proc Natl Acad Sci USA. 2010;107:13129–13134. doi: 10.1073/pnas.1002285107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Beliakoff J, Lee J, Ueno H, Aiyer A, Weissman IL, Barsh GS, Cardiff RD, Sun Z. The PIAS-like protein Zimp10 is essential for embryonic viability and proper vascular development. Mol Cell Biol. 2008;28:282–292. doi: 10.1128/MCB.00771-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Betancourt J, Katzman S, Chen B. Nuclear factor one B regulates neural stem cell differentiation and axonal projection of corticofugal neurons. J Comp Neurol. 2014;522(1):Spc1. doi: 10.1002/cne.23373. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bloom AJ, Miller BR, Sanes JR, DiAntonio A. The requirement for Phr1 in CNS axon tract formation reveals the corticostriatal boundary as a choice point for cortical axons. Genes Dev. 2007;21:2593–2606. doi: 10.1101/gad.1592107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bortone DS, Olsen SR, Scanziani M. Translaminar inhibitory cells recruited by layer 6 corticothalamic neurons suppress visual cortex. Neuron. 2014;82:474–485. doi: 10.1016/j.neuron.2014.02.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Briggs F. Organizing principles of cortical layer 6. Front Neural Circuits. 2010;12(4):3. doi: 10.3389/neuro.04.003.2010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cederquist GY, Azim E, Shnider SJ, Padmanabhan H, Macklis JD. Lmo4 establishes rostral motor cortex projection neuron subtype diversity. J Neurosci. 2013;33:6321–6332. doi: 10.1523/JNEUROSCI.5140-12.2013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen B, Schaevitz LR, McConnell SK. Fezl regulates the differentiation and axon targeting of layer 5 subcortical projection neurons in cerebral cortex. Proc Natl Acad Sci USA. 2005;102:17184–17189. doi: 10.1073/pnas.0508732102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen JG, Rasin MR, Kwan KY, Sestan N. Zfp312 is required for subcortical axonal projections and dendritic morphology of deep-layer pyramidal neurons of the cerebral cortex. Proc Natl Acad Sci USA. 2005;102:17792–17797. doi: 10.1073/pnas.0509032102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen B, Wang SS, Hattox AM, Rayburn H, Nelson SB, McConnell SK. The Fezf2-Tbr1 genetic pathway regulates the fate choice of subcortical projection neurons in the developing cerebral cortex. Proc Natl Acad Sci USA. 2008;105:11382–11387. doi: 10.1073/pnas.0804918105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Christoforou N, Chellappan M, Adler AF, Kirkton RD, Wu T, Addis RC, Bursac N, Leong KW. Transcription factors MYOCD, SRF, Mesp1 and SMARCD3 enhance the cardio-inducing effect of GATA4, TBX5, and MEF2C during direct cellular reprogramming. PLoS One. 2013;8:e63577. doi: 10.1371/journal.pone.0063577. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cubadda Y, Heitzler P, Ray RP, Bourouis M, Ramain P, Gelbart W, Simpson P, Haenlin M. U-shaped encodes a zinc finger protein that regulates the proneural genes achaete and scute during the formation of bristles in Drosophila. Genes Dev. 1997;11:3083–3095. doi: 10.1101/gad.11.22.3083. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Greig LC, Woodworth MB, Galazo MJ, Padmanabhan H, Macklis JD. Molecular logic of neocortical projection neuron specification, development, and diversity. Nat Rev Neurosci. 2013;14:755–769. doi: 10.1038/nrn3586. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Greig LC, Woodworth MB, Greppi C, Macklis JD. Ctip1 controls acquisition of sensory area identity and establishment of sensory input fields in the developing neocortex. Neuron. 2016 doi: 10.1016/j.neuron.2016.03.008. http://dx.doi.org/10.1016/j.neuron.2016.03.008. [DOI] [PMC free article] [PubMed]
- Ferland RJ, Cherry TJ, Preware PO, Morrisey EE, Walsh CA. Characterization of Foxp2 and Foxp1 mRNA and protein in the developing and mature brain. J Comp Neurol. 2003;460:266–279. doi: 10.1002/cne.10654. [DOI] [PubMed] [Google Scholar]
- Guillery RW, Sherman SM. Thalamic relay functions and their role in corticocortical communication: generalizations from the visual system. Neuron. 2002;33:163– 175. doi: 10.1016/s0896-6273(01)00582-7. [DOI] [PubMed] [Google Scholar]
- Han W, Kwan KY, Shim S, Lam MMS, Shin Y, Xu X, Zhu Y, Li M, Sestan N. TBR1 directly represses Fezf2 to control the laminar origin and development of the corticospinal tract. Proc Natl Acad Sci USA. 2011;108:3041–3046. doi: 10.1073/pnas.1016723108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harb K, Magrinelli E, Nicolas CS, Lukianets N, Frangeul L, Pietri M, Sun T, Sandoz G, Grammont F, Jabaudon D, Studer M, Alfano C. Area-specific development of distinct projection neuron subclasses is regulated by postnatal epigenetic modifications. Elife. 2016 Jan 27;:5. doi: 10.7554/eLife.09531. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hsueh HP. The role of the MAGUK protein CASK in neural development and synaptic function. Curr Med Chem. 2006;13:1915–1927. doi: 10.2174/092986706777585040. [DOI] [PubMed] [Google Scholar]
- Huggins GS, Bacani CJ, Boltax J, Aikawai R, Leiden JM. Friend of GATA 2 Physically Interacts with Chicken Ovalbumin Upstream Promoter-TF2 (COUP- TF2) and COUP-TF3 and Represses COUP-TF2 dependent Activation of the Atrial Natriuretic Factor Promoter. J Biol Chem. 2001;276:28029–28036. doi: 10.1074/jbc.M103577200. [DOI] [PubMed] [Google Scholar]
- Konopka G, Bomar JM, Winden K, Coppola G, Jonsson ZO, Gao F, Peng S, Preuss TM, Wohlschlegel JA, Geschwind DH. Human-specific transcriptional regulation of CNS development genes by FOXP2. Nature. 2009;462:213–217. doi: 10.1038/nature08549. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kuo TY, Hong CJ, Chien HL, Hsueh YP. X-linked mental retardation gene CASK interacts with Bcl11A/CTIP1 and regulates axon branching and outgrowth. J Neurosci Res. 2010;88:2364–2373. doi: 10.1002/jnr.22407. [DOI] [PubMed] [Google Scholar]
- Kwan KY, Lam MMS, Krsnik Z, Kawasawa YI, Lefebvre V, Sestan N. SOX5 postmitotically regulates migration, postmigratory differentiation, and projections of subplate and deep-layer neocortical neurons. Proc Natl Acad Sci USA. 2008;105:16021–16026. doi: 10.1073/pnas.0806791105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kwan K. Transcriptional dysregulation of neocortical circuit assembly in ASD. Int Rev Neurobiol. 2013;113:167–205. doi: 10.1016/B978-0-12-418700-9.00006-X. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lai T, Jabaudon D, Molyneaux BJ, Azim E, Arlotta P, Menezes JRL, Macklis JD. SOX5 Controls the Sequential Generation of Distinct Corticofugal Neuron Subtypes. Neuron. 2008;57:232–247. doi: 10.1016/j.neuron.2007.12.023. [DOI] [PubMed] [Google Scholar]
- Lin AC, Roche AE, Wilk J, Svensson EC. The N-termini of Friend of GATA (FOG) proteins define a novel transcriptional repression motif and a superfamily of transcriptional repressors. J Biol Chem. 2004;279:55017–55023. doi: 10.1074/jbc.M411240200. [DOI] [PubMed] [Google Scholar]
- Manuylov NL, Smagulova FO, Tevosian SG. Fog2 excision in mice leads to premature mammary gland involution and reduced Esr1 gene expression. Oncogene. 2007;26:5204–5213. doi: 10.1038/sj.onc.1210333. [DOI] [PubMed] [Google Scholar]
- McKenna WL, Betancourt J, Larkin KA, Abrams B, Guo C, Rubenstein JL, Chen B. Tbr1 and Fezf2 regulate alternate corticofugal neuronal identities during neocortical development. J Neurosci. 2011;31:549–564. doi: 10.1523/JNEUROSCI.4131-10.2011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Minegishi N, Ohta J, Yamagiwa H, Suzuki N, Kawauchi S, Zhou Y, Takahashi S, Hayashi N, Engel JD, Yamamoto M. The mouse GATA-2 gene is expressed in the para-aortic splanchnopleura and aorta-gonads and mesonephros region. Blood. 1999;93:4196–4207. [PubMed] [Google Scholar]
- Mitchell KJ. The genetics of neurodevelopmental disease. Curr Op Neurobiol. 2011;21:197–203. doi: 10.1016/j.conb.2010.08.009. [DOI] [PubMed] [Google Scholar]
- Molyneaux BJ, Arlotta P, Hirata T, Hibi M, Macklis JD. Fezl is required for the birth and specification of corticospinal motor neurons. Neuron. 2005;47:817–831. doi: 10.1016/j.neuron.2005.08.030. [DOI] [PubMed] [Google Scholar]
- Molyneaux BJ, Arlotta P, Fame RM, MacDonald JL, MacQuarrie KL, Macklis JD. Novel subtype-specific genes identify distinct subpopulations of callosal projection neurons. J Neurosci. 2009;29:12343–12354. doi: 10.1523/JNEUROSCI.6108-08.2009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Olsen SR, Bortone DS, Adesnik H, Scanziani M. Gain control by layer six in cortical circuits of vision. Nature. 2012;483:47–52. doi: 10.1038/nature10835. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Paganoni S, Ferreira A. Neurite extension in central neurons: a novel role for the receptor tyrosine kinases Ror1 and Ror2. J Cell Sci. 2005;118:433–446. doi: 10.1242/jcs.01622. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Perdomo J, Jiang XM, Carter DR, Khachigian LM, Chong BH. SUMOylation regulates the transcriptional repression activity of FOG-2 and its association with GATA-4. PLoS One. 2012;7:e50637. doi: 10.1371/journal.pone.0050637. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Piran S, Bassett AS, Grewal J, Swaby J, Morel C, Oechslin EN, Redington AN, Peter P, Liu PP, Silversides CK. Patterns of cardiac and extracardiac anomalies in adults with tetralogy of Fallot. Am Heart J. 2011;161:131–137. doi: 10.1016/j.ahj.2010.09.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reimers-Kipping S, Hevers W, Pääbo S, Enard W. Humanized Foxp2 specifically affects cortico-basal ganglia circuits. Neuroscience. 2011;175:75–84. doi: 10.1016/j.neuroscience.2010.11.042. [DOI] [PubMed] [Google Scholar]
- Sengar AS, Ellegood J, Yiu AP, Wang H, Wang W, Juneja SC, Lerch JP, Josselyn SA, Henkelman RM, Salter MW, Egan SE. Vertebrate intersectin1 is repurposed to facilitate cortical midline connectivity and higher order cognition. J Neurosci. 2013;33:4055–4065. doi: 10.1523/JNEUROSCI.4428-12.2013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Srinivasan K, Leone DP, Bateson RK, Dobreva G, Kohwi Y, Kohwi-Shigematsu T, Grosschedl R, McConnell SK. A network of genetic repression and derepression specifies projection fates in the developing neocortex. Proc Natl Acad Sci U S A. 2012;109:19071–19078. doi: 10.1073/pnas.1216793109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tevosian SG, Deconinck AE, Cantor AB, Rieff HI, Fujiwara Y, Corfas G, Orkin SH. FOG-2: A novel GATA-family cofactor related to multitype zinc-finger proteins Friend of GATA-1 and U-shaped. Proc Natl Acad Sci USA. 1999;96:950–955. doi: 10.1073/pnas.96.3.950. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tevosian SG, Deconinck AE, Tanaka M, Schinke M, Litovsky SH, Izumo S, Fujiwara Y, Orkin SH. FOG2, a cofactor for GATA transcription factors, is essential for heart morphogenesis and development of coronary vessels from epicardium. Cell. 2000;101:729–739. doi: 10.1016/s0092-8674(00)80885-5. [DOI] [PubMed] [Google Scholar]
- Tevosian SG, Albrecht KH, Crispino JD, Fujiwara Y, Eicher EM, Orkin SH. Gonadal differentiation, sex determination and normal Sry expression in mice require direct interaction between transcription partners GATA4 and FOG2. Development. 2002;129:4627–4634. doi: 10.1242/dev.129.19.4627. [DOI] [PubMed] [Google Scholar]
- Thierry G, Pichon O, Briand A, Damien Poulain D, Sznajer Y, David A, Le Caignec C. Autosomal insertional translocation mimicking an X-linked mode of inheritance. European J Med Genetics. 2013;56:46–49. doi: 10.1016/j.ejmg.2012.10.006. [DOI] [PubMed] [Google Scholar]
- Tripodi M, Filosa A, Armentano M, Studer M. The COUP-TF nuclear receptors regulate cell migration in the mammalian basal forebrain. Development. 2004;131:6119–6129. doi: 10.1242/dev.01530. [DOI] [PubMed] [Google Scholar]
- Wat MJ, Veenma D, Hogue J, Holder AM, Yu Z, Wat J, Hanchard N, Shchelochkov OA, Fernandes CJ, Johnson A, Kevin P, Lally KP, Slavotinek A, Danhaive O, Schaible T. Genomic alterations that contribute to the development of isolated and non-isolated congenital diaphragmatic hernia. J Med Genet. 2011;48:299–307. doi: 10.1136/jmg.2011.089680. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wilkinson-White L, Gamsjaeger R, Dastmalchi S, Wienert B, Stokes PH, Crossley M, Mackay JP, Matthews JM. Structural basis of simultaneous recruitment of the transcriptional regulators LMO2 and FOG1/ZFPM1 by the transcription factor GATA1. Proc Natl Acad Sci USA. 2011;108:14443–14448. doi: 10.1073/pnas.1105898108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Woodworth MB, Greig LC, Liu KX, Ippolito GC, Tucker HO, Macklis JD. Ctip1 regulates the balance between specification of distinct projection neuron subtypes in deep cortical layers. Cell Reports. 2016 doi: 10.1016/j.celrep.2016.03.064. http://dx.doi.org/10.1016/j.celrep.2016.03.064. [DOI] [PMC free article] [PubMed]
- Yao J, Liu Y, Husain J, Lo R, Palaparti A, Henderson J, Stifani S. Combinatorial expression patterns of individual TLE proteins during cell determination and differentiation suggest non-redundant functions for mammalian homologs of Drosophila Groucho. Develop Growth Differ. 1998;40:133–146. doi: 10.1046/j.1440-169x.1998.00003.x. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Figure S1. (related to Figure 1) Genes identified by microarray analysis are expressed in the cortical plate and layer VI.
In situ hybridization or immunocytochemistry showing expression in layer VI for all genes selected from Figure 1 (except genes shown in Figure 2). For each gene, the temporal course of gene expression from microarray analysis by CThPN (green), CSMN (red), and CPN (blue) are shown in the same panel. Expression is shown at E16 or postnatal stages (P0, P1, or P3). For genes expressed by CThPN subpopulations (Zmiz1, Cask, Dmrt2, Phr1, Zmynd11), arrowheads indicate restricted expression. Immuncytochemistry for CASK detects CThPN in lower layer VI, and CThPN axons in the internal capsule (white arrowheads). Scale bars, 250 μm (E16), 150 μm (P0) and 500 μm (P1, P3).
Figure S2. (related to Figure 3) Fog2 is expressed by CThPN across cortical areas.
(A–C) Fog2 is strongly expressed in layer VI by CThPN, and by other neuron subtypes in other forebrain regions. (D–I) In the cortex, FOG2 is excluded from high level CTIP2- expressing neurons (SCPN) (D–F), and SATB2-expressing neurons (CPN, arrows) (G–I). (J–N) FOG2 expression is CThPN specific, compared to TBR1, also expressed in layer VI CPN (white arrowheads in L–N). Scale bars, 500 μm (A–C), 100 μm (D–F, J), 20 μm (G–I) and 10 μm (L–N).
Figure S3. (related to Figure 4) Cortical morphology and expression of widely used layer VI markers are not affected by the absence of Fog2 function in cortex. (A–B) Fog2 cKO mice show absence of Fog2 mRNA specifically in the neocortex (layer VI, black arrowheads), but show normal expression in other forebrain regions. (C–D) Nissl staining of cortex in wild-type (C) and Fog2 cKO (D) mice at P7 reveals no differences in laminar patterning. (E–L) IHC for TBR1 (E, I), TLE4 (F, J), FOXP2 (G, K), and ISH for Fezf2 (H, L) show no difference in expression of these markers between WT and Fog2 cKO cortices. (M–R) FluoroGold injections in thalamic nuclei in WT and Fog2 cKO brains: (M, P) Mediodorsal nucleus (MD); (N, Q) Ventral anterior/Ventro lateral nucleus (VA/VL); and (O, R) Ventrobasal complex (VB). Scale bar, 50 μm (E–K) and 500 μm (M–R).
Figure S4. (related to Figure 5) In the absence of Fog2 function, SCPN are born and migrate normally. (A–C) Quantification of E13.5 BrdU birthdated neurons at P6 across the thickness of motor cortex. There is no difference between WT and Fog2 cKO cortex in the number or distribution of BrdU+ neurons at P6 that were labeled at E13.5, indicating that SCPN generation and migration are not altered in Fog2 cKO mice. Scale bar, 100 μm (A–B). Data are represented as mean ± SEM.
Figure S5. (related to Figure 6) Axonal projections to the cerebral peduncle are normal in the absence of Fog2 function. (A–E) SCPN are correctly located in layer V, and not in layer VI, in motor cortex of Fog2 cKO mice. (A) Schematic of experimental approach. (B, C) Low magnification images of the area analyzed. (D–E) High magnification of area boxed in B–C. Scale bar, 60 μm (D–E).
Figure S6. (related to Figure 7) Fog2 is mis-expressed exclusively in Cre-electroporated neurons in CTV-Fog2 embryos (CAG-floxed-Stop-Fog2-IRES-Gfp). (A–D′) Cre and tdTomato co-electroporation in CTV-Fog2 embryos at E13.5 results in Fog2 mis- expression in layer V and upper-layers, only in Cre-recombined neurons (GFP+, FOG2+ detected by immunolabeling; white arrowheads in A′–D′). FOG2 is not expressed in tdTomato-only electroporated neurons (open arrowheads, A′–D′). (A′–D′) High magnification of area boxed in A–D. Scale bars, 50 μm (A–D) and 25 μm (A′–D′).
Figure S7. (related to Figure 8) COUPTF1, COUPTF2, GATA2, and GATA4 expression in the developing cortex. Immunocytochemistry for FOG2, COUPTF1, and GATA2 shows expression of these proteins in the preplate of developing cortex at E12.5 (A, B, D), and in the cortical plate at E15.5 (E, F, H). Higher magnification images of the boxed areas corresponding to developing motor cortex are shown in (A′– H′). Immuncytochemistry for COUPTF2 shows expression primarily by progenitors and meningeal cells, but not in the preplate at E12.5 (C-C′), nor in the dorsal cortical plate at E15.5 (G-G′). Immuncytochemistry for GATA4 shows expression at E15.5 in a medio-lateral gradient, with highest expression in the medial and dorsal cortical plate, but little or no expression the lateral cortical plate (I-I′). No expression was detected at earlier time points (not shown). Scale bars, 20 μm (A′–I′).
Supplemental Table 1. (related to Figure 1) Summary of Gene Ontology terms for biological functions over-represented among CThPN-genes.
Supplemental Table 2. (related to Figure 1 and Figure 2) cDNA clones for riboprobes.


