Figure 2. Commonly mutated spliceosomal proteins and their associations with specific cancer types.
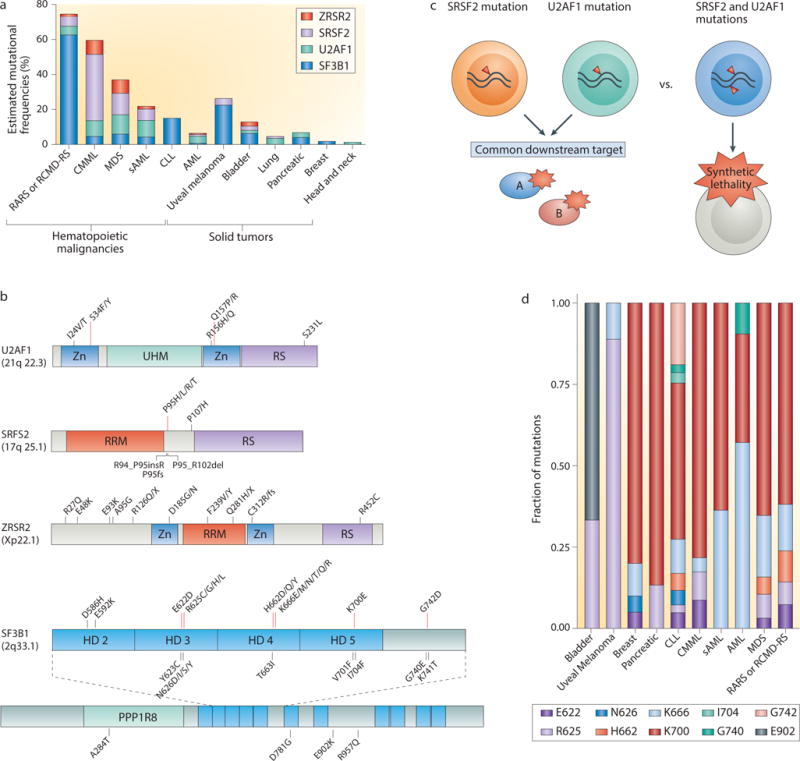
(a) The incidence and cancer distribution of the most frequently mutated spliceosomal genes are shown. (b) The spectrum of mutations that have been identified in U2AF1 (U2 small nuclear RNA auxiliary factor 1), SRSF2 (serine/arginine-rich splicing factor 2), ZRSR2 (zinc finger, RNA-binding motif and serine/arginine-rich 2) and SF3B1 (splicing factor 3B, subunit 1). Mutations shown in bold text occur at hotspots; other illustrated mutations are recurrent but rare. As ZRSR2 mutations do not occur at hotspots, very rare or private mutations are shown as examples. (c) Schematic illustrating the potential reasons that spliceosomal gene mutations are mutually exclusive with one another: they either converge on a common downstream target or result in synthetic lethality. (d) The frequency of specific mutations in SF3B1 across various histological subtypes of cancer. AML, acute myeloid leukemia; CLL, chronic lymphocytic leukemia; CMML, chronic myelomonocytic leukemia; HD, HEAT domain; PPP1R8, binding site for protein phosphatase 1 regulatory subunit 8; RARS, refractory anemia with ringed sideroblasts; RCMD-RS, refractory cytopenia with multilineage dysplasia and ringed sideroblasts; RRM, RNA recognition motif; RS, arginine/serine-rich domain; sAML, secondary AML; UHM, U2AF homology motif; ZN, zinc finger domain.
