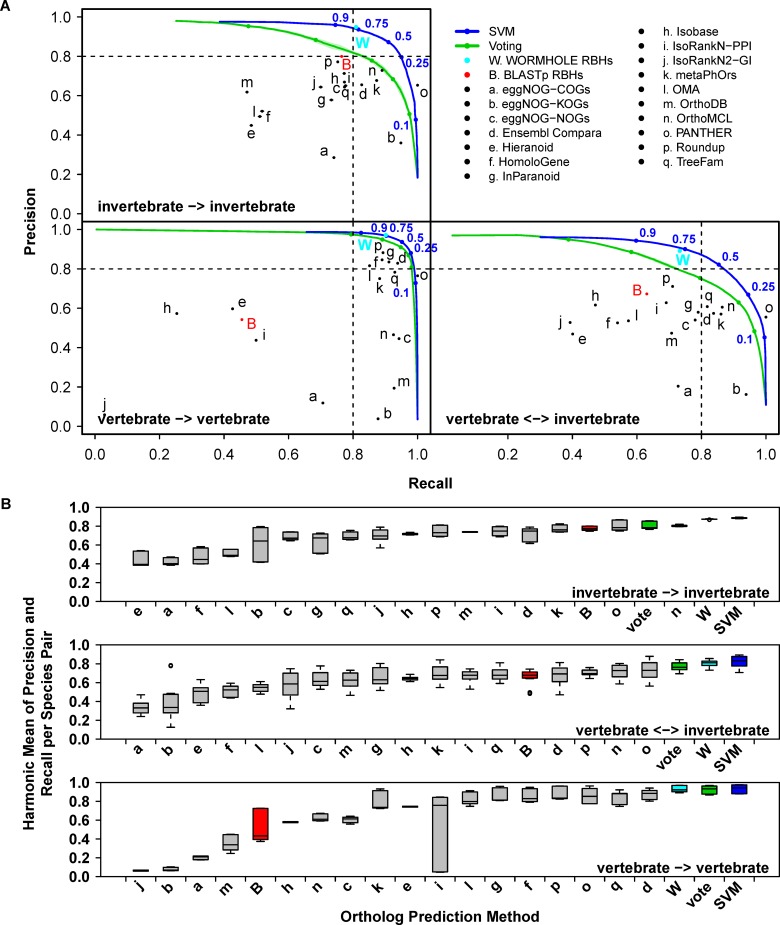
Fig 2. WORMHOLE SVMs improve prediction of PANTHER LDOs over constituent algorithms and voting to a degree dependent on the evolutionary separation of the compared species.
(A) Precision-recall performance charts for PANTHER LDO predictions made between vertebrate and invertebrate species separated into categories based on evolutionary distance. Points or lines represent the mean performance of the 17 constituent algorithms (black), BLASTp reciprocal best hits (RBHs) (red), voting (green), WORMHOLE SVMs (blue), or WORMHOLE RBHs (cyan) at predicting PANTHER LDOs across the 10-folds of the outer cross-validation (see Materials and Methods). WORMHOLE RBHs are reciprocal best hits selected based on the WORMHOLE Score and are introduced later in the Results section. Error bars and colored regions represent standard error of mean for precision and recall across folds (due to the large number of gene pairs, error bars and regions are small and fall within the width of the point or line in most cases). Lines are generated by sampling the complete range of possible threshold values for each confidence score type. Color-matched points indicate the performance for specified threshold values (blue numbers) on each line. (B) Box and whisker plot representing the harmonic mean of precision and recall for each of the 17 constituent WORMHOLE algorithms, voting, BLASTp RBHs, WORMHOLE SVMs, and WORMHOLE RBHs when predicting PANTHER LDOs for each pair of query and target species. Ortholog prediction methods are ordered by median harmonic mean. For voting, SVMs, and WORMHOLE RBHs, values represent the maximum harmonic mean for each pair of query and target species (WORMHOLE Score ≥ 0.5).