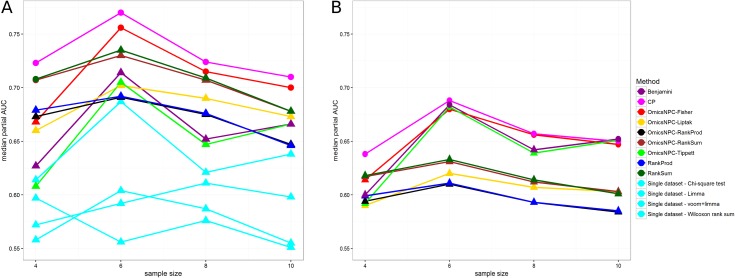
Fig 3. Median pAUCs in relation to the number of samples in the different modalities scenario.
Each line corresponds to a specific method. The x axis represents the number of samples per group used in the simulations, and y axis the median pAUCs. Circles suggest that the observed differences from the best method were not statistically significant at level ≤ 0.05. A) Only genes which exhibited the same behaviour across datasets were taken into account. The integrative analysis yielded better results regardless the employed method. CP and omicsNPCFisher were the best method in most cases. B) All genes were considered in this analysis. Again, omicsNPCFisher and CP achieved better performance than other methods in most cases; the Benjamini method was also statistically indistinguishable from the best performing method when the sample size was 6 or 10.