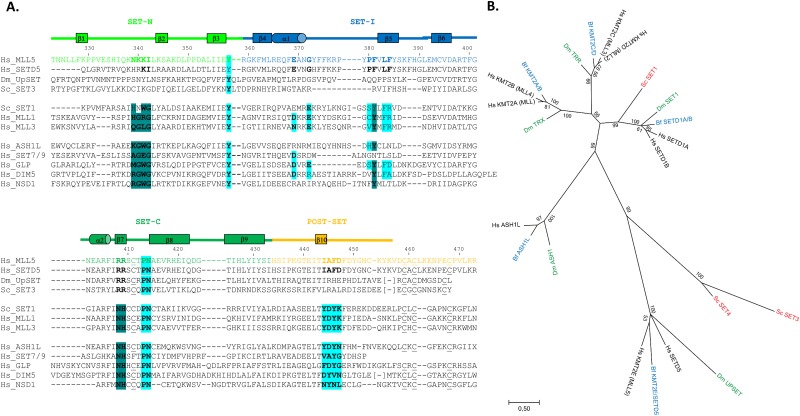
Fig 1. MLL5 SET domain lacks important residues for methyltransferase activity.
(A). Sequence alignment of the human MLL5 SET domain with SET domains from other methyltransferases (human SETD5, Drosophila UPSET, yeast SET3, yeast SET1, human MLL1, human MLL3, human ASH1L, human SET7/9, human GLP, human DIM5 and human NSD1). Structural alignment was calculated using the Dali server [56] when structure of the SET domain were available. Protein subdomains, indicated above the sequences, are based on the human MLL1 SET domain [14]. Secondary structure elements, derived from the structure of the MLL5 SET domain, are also shown above the sequences. Residues usually important for cofactor binding (dark blue) and for substrate binding and maintenance in the active center (cyan) are highlighted. Conserved cysteines from the zinc-binding cage are underlined. (B). Phylogenetic tree of methyltransferases related to MLL5 using sequences from humans (Homo sapiens) (Hs, black), the cephalochordate amphioxus (Branchiostoma floridae) (Bf, blue), the fruit fly (Drosophila melanogaster) (Dm, green) and the yeast (Saccharomyces cerevisiae) (Sc, red). The tree resulting from the Maximum Likelihood (ML) analysis is shown and branch lengths are representative of sequence substitution rates. Branch support is indicated as Bootstrap percentages (ranging from 0 to 100) and as posterior probabilities (ranging from 0 to 1) obtained from the Bayesian Inference (BI) analysis (provided in parentheses at each node). “(—)” indicates that the branching patterns of the ML and BI analyses diverged at this node and that a posterior probability score is thus unavailable.