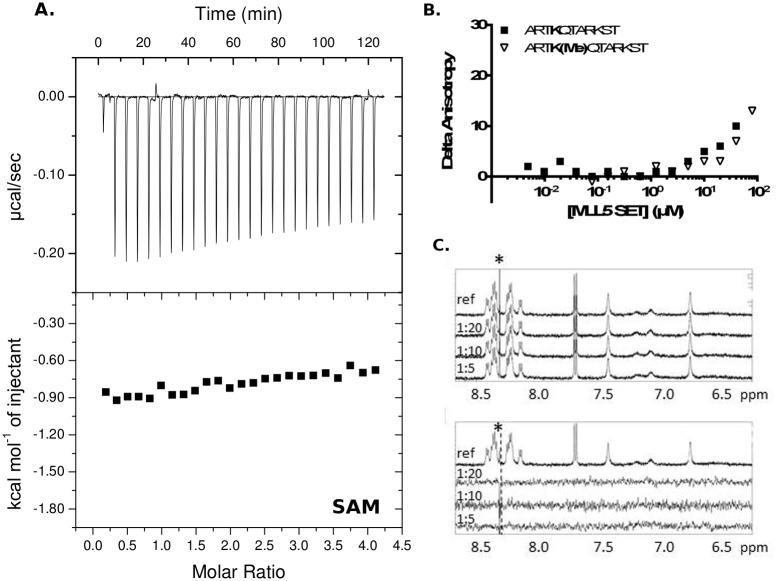
Fig 2. Absence of cofactor and substrate (H3K4 peptide) binding to the MLL5 SET domain.
(A). Isothermal titration calorimetry shows that the MLL5 SET domain in combination with the N-terminal flanking helix and the POSTSET does not bind the cofactor S-adenosyl methionine (SAM). Results were the same with the fragment SET-POSTSET. (B). Fluorescence anisotropy shows that the MLL5 SET domain (SET-POSTSET construct) does not bind either the non-methylated or mono-methylated H3K4 peptide. (C). 1H NMR spectra of SET-POSTSET construct and un-methylated H3K4 peptide. Upper panel: Expansion of 1H NMR spectra of un-methylated H3K4 peptide (top) at 400 μM, and in presence of different ratios protein/peptide (1:20, 1:10, 1:5). Lower panel: Expansion of 1H (top) and saturation transfer difference (STD) spectra of un-methylated H3K4 peptide at 400 μM in presence of different ratios of protein. In all spectra, the fine peak labeled with an asterisk (*), if present, corresponds to an impurity, whose resonance is imperfectly subtracted in STD on- and off-resonance experiments.