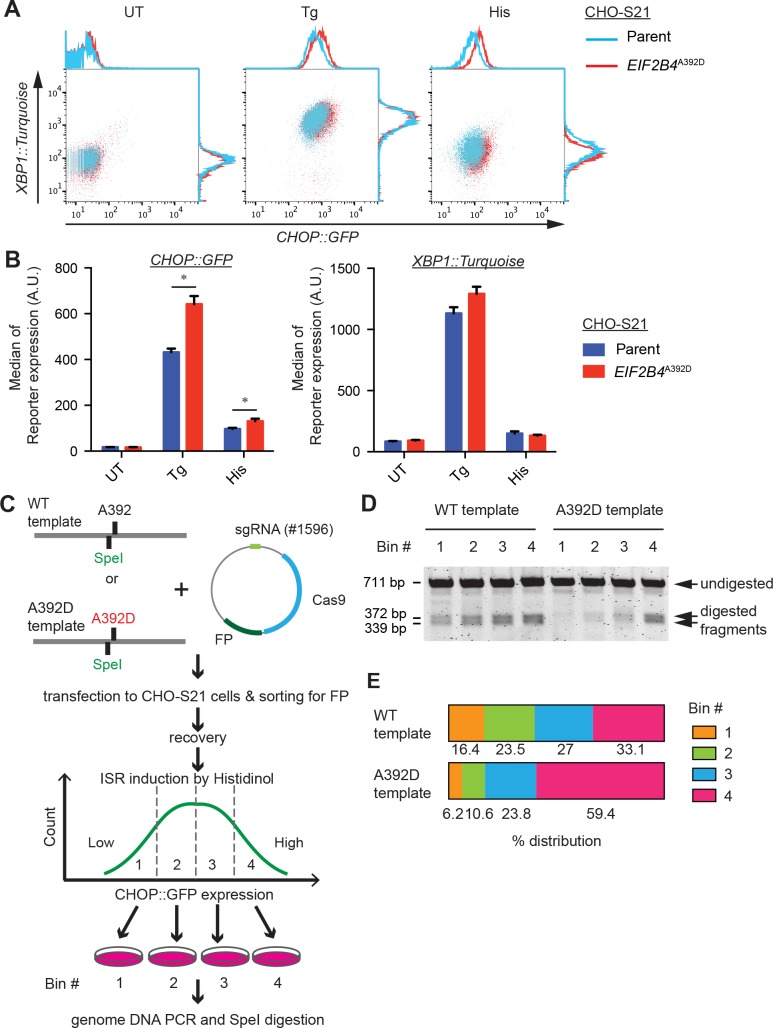
Fig 2. Heightened ISR in EIF2B4A392D cells.
(A) Flow cytometry analysis of CHOP::GFP and XBP1::Turquoise dual reporter-containing parental CHO-S21 and EIF2B4A392D mutant cells. The cells were untreated (UT) or stimulated with 250 nM thapsigargin (Tg) or 0.5 mM histidinol (His) for 24 hours. Note the enhanced response of the CHOP::GFP ISR reporter. (B) Bar diagram of the median ± S.D. of the reporter gene activity from experiments as shown in “A”. N = 3, *P = 0.0057 for Tg, *P = 0.037 for His, Unpaired t test. (C) Experimental design for tracking EIF2B4A392D mutations. A fluorescent protein-marked sgRNA/Cas9 plasmid targeting EIF2B4 and a wildtype or EIF2B4A392D mutant repair template marked by a silent SpeI mutation were co-transfected into CHO-S21 cells. Transfected cells (selected by FACS), were treated with histidinol and divided into four bins (Bin #1 to #4) by level of CHOP::GFP expression. After recovery, genomic DNA was isolated from cells in each bin and the targeted region of EIF2B4 was amplified by PCR and digested with SpeI to reveal frequency of targeting by either repair template. (D) PCR fragments digested with SpeI from genomic DNA of the indicated bins, visualized on an agarose gel. Shown is an image of a representative experiment reproduced twice. (E) Plot of the distribution of SpeI digested fragments in the four bins of transduced cells from the experiment in “D”. The band intensities of the digested fragments (reporting on recombination of the wildtype or mutant repair template) were normalized to total PCR product intensity and the distribution of the relative frequency of recombination in the different bins was plotted. Note the enrichment for recombination of the EIF2B4A392D mutant repair template in the ISRHigh bin.