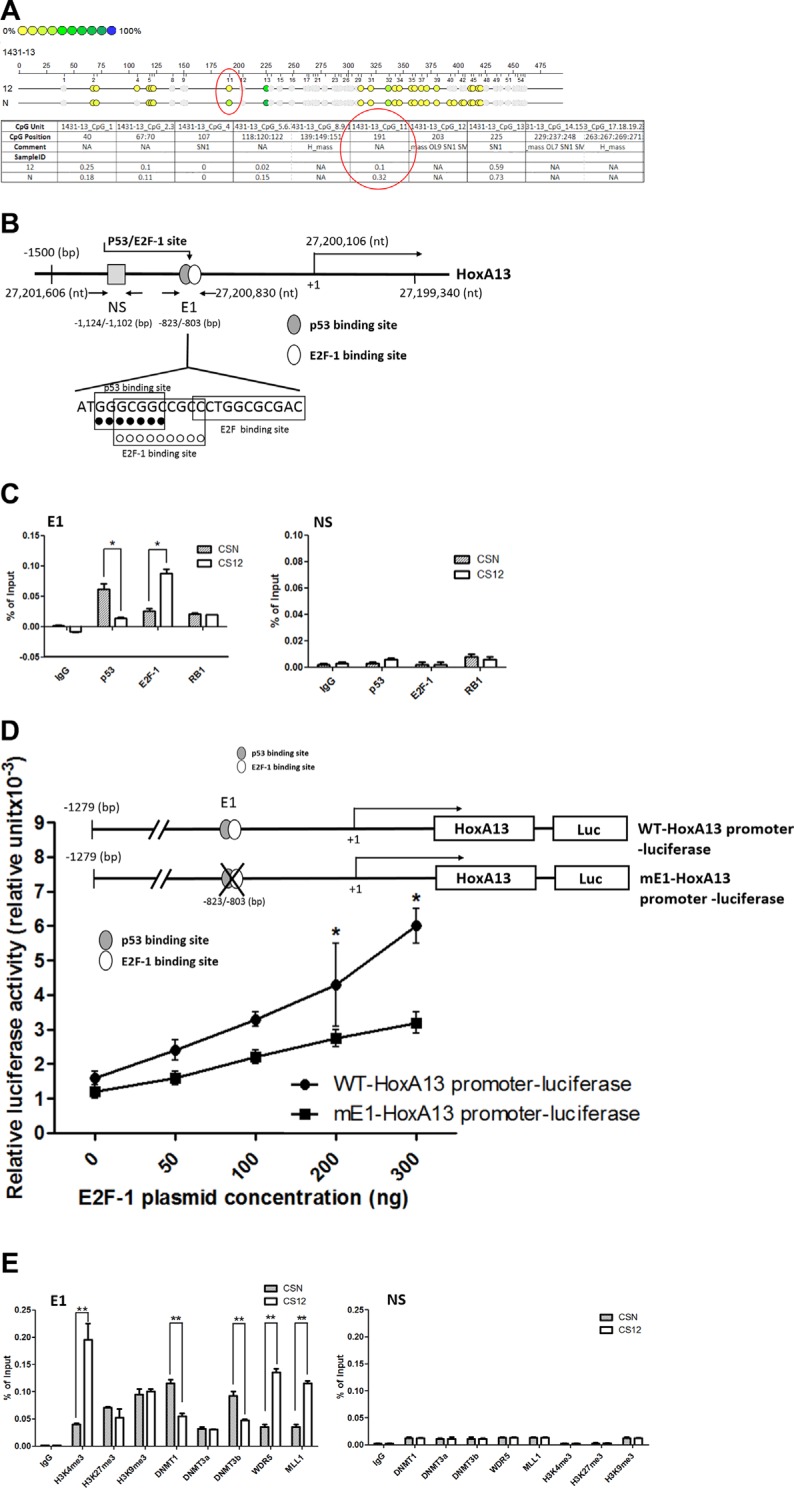
Figure 4. DNA methylation of E1 site in CpG islands of HoxA13 promoter.
(A) DNA methylation analysis of HoxA13 promoter. The relative extent of DNA methylation is indicated as intensity; complete methylation with a value of 1.0 is shown as green and hypomethylation is indicated by yellow dots. The red circle shows significant difference in DNA methylation at CpG 11 (1,431–13 segment) of the HoxA13 promoter. N: CSN cells; 12; CS12 cells. NA indicates not added. (B) Schematic representation of the promoter region of HoxA13 gene. The differential methylated CpG 11 site was found to be the p53/E2F-1 binding site (E1 site; −823 to −803 bp). A nonspecific site (NS; gray square) for ChIP-qPCR was assigned at −1,124 to −1,102 (bp). +1 indicates the transcriptional start site. (C) ChIP–qPCR analysis of p53, E2F-1, RB-1, and IgG (negative control) was performed in CSN and CS12 cells as described in the Materials and Methods. Input DNA (1/20-fold) was also analyzed. (D) Luciferase-linked wild type (WT) or mE1 mutant-promoter, control pGL4, and various amounts (0, 50, 100, 200, and 300 ng) of pCMV-SPORT6-E2F-1 were transfected into CS12 cells, and luciferase activity was measured as described in the Materials and Methods. (E) ChIP–qPCR analysis using antibodies against DNA methyltransferase family members, methylated histones, and the WDR5–MLL complex were performed in CSN and CS12 cells. Input DNA (1/20-fold) was also analyzed. The data C-E are presented as mean ± SEM (two-tailed Student t test; *p < 0.05, **p < 0.01).