Figure 4.
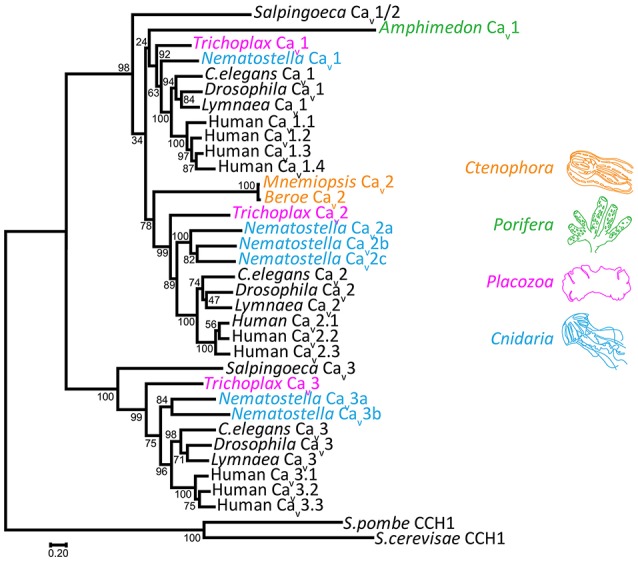
Maximum likelihood protein phylogeny of select Cav channels from animals, rooted with Cav channel homologs from fungi. Inference was achieved using MUSCLE protein alignment with MEGA7, followed by alignment trimming with TrimAL. Evolutionary models for maximum likelihood phylogenetic inference were tested with MEGA7, indicating that the LG matrix with gamma frequencies was the best fit using both corrected Akaike's Information Criterion and Bayesian Information Criterion. Node support values from 500 bootstrap replicates are indicated. GenBank accession numbers: Salpingoeca Cav1/Cav2: XP_004989719; Amphimedon Cav1/Cav2: XP_003383036; Trichoplax Cav1: XP_002108930; Trichoplax Cav2: XP_002109775; Trichoplax Cav3: KJ466205; C.elegans Cav1 (egl-19): NP_001023079; C.elegans Cav2 (unc-2): NP_001123176; C.elegans Cav3 (cca-1): CCD68017; Drosophila Cav1 (α1-D): AAF53504; Drosophila Cav2 (cacophony): AFH07350; Drosophila Cav3 (Ca-α1T): ABW09342; Lymnaea Cav1: AAO83839; Lymnaea Cav2: AAO83841; Lymnaea Cav3: AAO83843; human Cav1.1: NP_000060.2; human Cav1.2: AAI46847.1; human Cav1.3: NP_001122312.1; human Cav1.4: NP_005174.2; human Cav2.1: O00555.2; human Cav2.2: NP_000709; human Cav2.3: NP_001192222.1; human Cav3.1: NP_061496; human Cav3.2: NP_066921; human Cav3.3: NP_066919; Mnemiopsis Cav2: AEF59085; S.cerevisae CCH1: P50077; S.pombe CCH1: NP_593894.1. Other accession numbers: Nematostella Cav1: JGI-Genome Portal protein ID 88037; Nematostella Cav2a, Cav2b, Cav2c, Cav3a, Cav3b: Transcript sequences from the sequenced transcriptome (Fredman et al., 2013) NVE4667, NVE18768, NVE1263, NVE5017, and NVE7616 respectively. Scale bar represents the number of amino acid substitutions per site along the sequence alignment.
