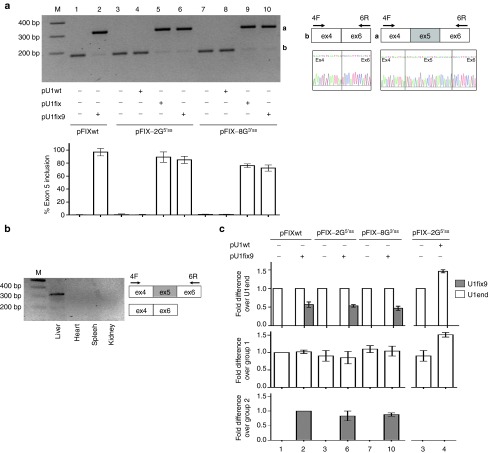
Figure 2.
Evaluation of the U1fix9-mediated rescue of hFIX mRNA splicing in mice. (a) Representative example of hFIX alternative splicing patterns in liver from BALB/c mice injected with the pFIX variants alone (−) or in combination (+) with a molar excess (1.5X) of the pU1wt, pU1fix or pU1fix9. The schematic representation of the normal and aberrant transcripts, and of primers used for the RT-PCR (arrows), is reported in the right panel. Amplified products were separated on 2% agarose gel. M, 100 bp molecular weight marker. The histograms report the relative percentage of correct transcripts in liver from mice injected at the different conditions (n = 4 for group 1–2, 4–5, 7–10; n = 5 and n = 7 for groups 3 and 6, respectively), and results are expressed as mean ± SD. (b) Representative example of hFIX splicing patterns in various organs of mice coinjected with pFIX-2G5′ss and pU1fix9. (c) Evaluation (middle and lower panels) and comparison (upper panel) of the expression of the endogenous (U1end, white histograms) and ExSpeU1 (U1fix9, gray histograms) by qPCR in liver from mice treated as in panel A. For qPCR, the results are expressed as mean ± SD from three mice for each condition. Values are normalized for the mouse GAPDH and are indicated as fold increase compared to the reference indicated in the Y-axis, set to 1. No transcripts were detected in samples without reverse transcriptase. ExSpeU1, Exon-specific U1snRNA; hFIX, human factor IX; qPCR, quantitative PCR; RT-PCR, reverse transcription polymerase chain reaction; SD, standard deviation.