Abstract
The reproducibility of gamma‐aminobutyric acid (GABA) quantification results, obtained with MRSI, was determined on a 3 T MR scanner in healthy adults. In this study, a spiral‐encoded, GABA‐edited, MEGA‐LASER MRSI sequence with real‐time motion–scanner‐instability corrections was applied for robust 3D mapping of neurotransmitters in the brain. In particular, the GABA+ (i.e. GABA plus macromolecule contamination) and Glx (i.e. glutamate plus glutamine contamination) signal was measured. This sequence enables 3D‐MRSI with about 3 cm3 nominal resolution in about 20 min. Since reliable quantification of GABA is challenging, the spatial distribution of the inter‐subject and intra‐subject variability of GABA+ and Glx levels was studied via test–retest assessment in 14 healthy volunteers (seven men–seven women).
For both inter‐subject and intra‐subject repeated measurement sessions a low coefficient of variation (CV) and a high intraclass correlation coefficient (ICC) were found for GABA+ and Glx ratios across all evaluated voxels (intra−/inter‐subject: GABA+ ratios, CV ~ 8%–ICC > 0.75; Glx ratios, CV ~ 6%–ICC > 0.70). The same was found in selected brain regions for Glx ratios versus GABA+ ratios (CV varied from about 5% versus about 8% in occipital and parietal regions, to about 8% versus about 10% in the frontal area, thalamus, and basal ganglia).
These results provide evidence that 3D mapping of GABA+ and Glx using the described methodology provides high reproducibility for application in clinical and neuroscientific studies.
Keywords: acquisition methods, MEGA editing, MRS and MRSI methods, neurotransmitters, normal brain, reproducibility
Abbreviations used
- CNS
central nervous system
- CRLB
Cramér–Rao lower bound
- CV
coefficient of variation
- FOV
field of view
- FWHM
full width at half maximum
- GABA
gamma‐aminobutyric acid
- GABA+
gamma‐aminobutyric acid + macromolecules
- Gln
glutamine
- Glu
glutamate
- Glx
glutamate + glutamine
- GM
gray matter
- GOIA
gradient offset independent adiabatic
- ICC
intraclass correlation coefficient
- LASER
localized adiabatic spin‐echo refocusing
- MEGA
Mescher–Garwood
- MM
macromolecule
- MPRAGE
magnetization‐prepared rapid gradient‐echo
- PRESS
point‐resolved spectroscopy
- SD
standard deviation
- SNR
signal‐to‐noise ratio
- SVS
single‐voxel spectroscopy
- tCr
creatine + phosphocreatine
- tNAA
N‐acetyl‐aspartate + N‐acetyl aspartyl glutamate
- VOI
volume of interest
- WM
white matter
1. INTRODUCTION
Brain neurotransmitters work as chemical exchange messengers between neurons and are responsible for both physiological and mental processes.1 Although glutamate (Glu) is the most abundant excitatory neurotransmitter in the central nervous system (CNS), with concentrations of 5–15 mM,1, 2 the in vivo levels of the major inhibitory neurotransmitter, gamma‐aminobutyric acid (GABA), reach only 1–2 mM.3 Both GABA and Glu are essential for the development and function of the healthy brain.4, 5 They play an important role as potential biomarkers for monitoring various neurological (e.g. Alzheimer's1, 6 and Parkinson's disease,1, 7 or epilepsy5, 8), psychiatric disorders (e.g. anxiety,9, 10 major depressive and bipolar disorder,1, 11 or schizophrenia12, 13), and other diseases of development.5, 10
The accurate quantification of these neurotransmitters, particularly GABA, is complicated, even when measured at 3 T.14 Due to J‐coupling effects, the MRS signal of Glu appears as a complicated triplet at 3.75 ppm and multiplets in the range of 2.4–2.04 ppm.15 Furthermore, glutamine (Gln) overlaps the Glu signal, and thus they are usually evaluated as the sum of both Glu and Gln (Glx).16 All the GABA proton multiplet signals that resonate at 1.88, 2.28, and 3.02 ppm 15 overlap entirely with other metabolite resonances, i.e. with creatine and phosphocreatine (tCr), with Glx, with N‐acetyl aspartate and N‐acetyl aspartyl glutamate (tNAA), and with macromolecules (MMs).4 Among dedicated MRS‐editing techniques for GABA quantification, the most popular is Mescher–Garwood (MEGA) editing.17, 18 MEGA is a J‐difference MRS‐editing technique that operates on two selective frequencies. During EDIT‐ON acquisition, the MEGA pulses influence metabolites that resonate at 1.9 ppm, as well as their J‐coupling partners (i.e. GABA at 1.9 and 3.02 ppm, Glx at 2.1 and 3.75 ppm, and NAA at 2.0 ppm).16, 18 During the EDIT‐OFF acquisition the metabolite resonances are not affected. The spectra from EDIT‐ON and EDIT‐OFF acquisitions are subtracted to obtain a difference spectrum.5 However, conventional J‐difference MRS does not separate the GABA signal entirely from contamination by co‐edited MM signals at 3.0 ppm, which are coupled to spins at 1.7 ppm.19. Thus, the derived GABA signal is usually labeled as GABA+.4
MEGA‐edited measurements are challenging, mainly due to B 0/B 1 + inhomogeneities, scanner instabilities, motion artifacts, and long scan times.18 Therefore, a spiral‐accelerated 3D‐MRSI sequence was developed with real‐time artifact corrections, which employs MEGA‐LASER editing, as described recently.20, 21 Localized adiabatic spin‐echo refocusing (LASER) selection eliminates the drawbacks of conventional RF pulses, including B 1 + changes due to different coil loadings and inhomogeneities, and maintains perfect 90° or 180° pulses. Other confounding factors could be B 1 − inhomogeneities, which can be corrected via appropriate coil combinations.22 Furthermore, when using gradient offset independent adiabatic (GOIA) pulses, LASER nearly eliminates chemical‐shift displacement errors and nonuniformity, and reduces power requirements (SAR, specific absorption rate).23, 24 In addition, spiral readout trajectories simultaneously acquire frequency in two spatial dimensions, thus enabling up to 50 times faster data sampling than conventional phase‐encoding approaches.23
For these reasons, MEGA‐LASER spiral 3D‐MRSI constitutes a promising tool for the robust acquisition of GABA+, Glx, and other important neurometabolites in vivo within large parts of the brain within one scan.20, 21 Despite the many advantages of this method, to date its reproducibility has not yet been determined.
The purpose of this study was therefore to measure the intra‐ and inter‐subject reproducibility of GABA+ and Glx in healthy subjects, and to compare with literature values previously determined using MEGA‐PRESS (point‐resolved spectroscopy).
2. EXPERIMENT
2.1. Subjects
Fourteen healthy volunteers (age 30 ± 5 years, seven males and seven females) were included in this study after institutional review board approval and obtaining written, informed consent. None of the subjects demonstrated pathological findings on MRI, had any known history of neurological disorders, or had any medical treatment. All volunteers were scanned twice, with a gap of 30 min or less, to minimize intra‐subject variability. During this gap, all participants were removed from the scanner.
2.2. Hardware
This study was performed on a 3 T whole‐body MR scanner (TIM Trio®, Siemens Healthcare, Erlangen, Germany) with 45 mT/m total gradient strength and 200 mT/m/s nominal slew. A body coil was used for transmission and a 32‐channel head coil was used for signal reception (Siemens Healthcare).
2.3. Measurement protocol and sequence parameters
For all measurements in volunteers, the following imaging protocol was followed. Auto‐align was used to ensure identical slice positioning in the brain between the two scan sessions, as well as similarity of positioning of the subjects.25 To ensure accurate volume of interest (VOI) placement, 3D T 1‐weighted anatomical reference images were acquired via a magnetization‐prepared rapid gradient‐echo (MPRAGE) sequence, with generalized auto‐calibrating partially parallel acquisition (GRAPPA) 3 and 2 min 27 s measurement time, and then these images were resliced in three orthogonal directions. An additional 3D, high‐resolution, T 1‐weighted MRI for tissue segmentation was measured, with a total acquisition time of 6 min 39 s. For spectroscopic measurements, a constant‐density, spiral‐encoded, 3D‐MRSI sequence with MEGA‐LASER editing was used. Real‐time correction was applied for rigid motion bias (i.e. translations and rotations) and correction of shim and center frequency changes.20, 21 All MRSI slices were placed parallel to the anterior commissure–posterior commissure line and covered the centrum semiovale and basal ganglia (Figure 1), with VOI = 80 × 85 × 60 mm3 and field of view (FOV) = 200 × 200 × 170 mm3. The acquired matrix size of 14 × 14 × 12 (i.e. ~3 cm3 nominal voxel size) was interpolated to a 16 × 16 × 16 matrix. During EDIT‐ON acquisition, the MEGA‐editing pulses (60 Hz Gaussian pulses of 14.8 ms duration) were set to 1.9 ppm, to edit the coupled 4CH2 triplet of GABA resonating at 3.02 ppm.18, 24 During EDIT‐OFF acquisition, the editing pulse was applied at 7.5 ppm, symmetrically around the water peak. The difference spectrum was derived on the scanner via subtraction of EDIT‐ON and EDIT‐OFF spectra (Figure 2). VOI selection via LASER and low‐power and wide‐bandwidth GOIA pulses enabled MEGA editing with an echo time of 68 ms.21 All adiabatic pulses were run with a B 1 + safety margin of 10% above the adiabatic threshold. For real‐time correction, volumetric, dual‐contrast, echo planar imaging‐based navigators that update B 0 shim, frequency, and head‐position changes for each pair of EDIT‐ON–OFF acquisitions were used (i.e. with a repetition time of 1.6 s, an update occurs every 3.2 s). To cover the x/y–k space and the spectral width properly, two temporal and two spatial interleaves were performed. For 3D‐MRSI, 16 acquisition‐weighted averages (for k‐space weighting a cosine shaped window function was used) and two‐step phase cycling (i.e., the phase of the ADC and the excitation pulse were toggled between 0° and 180°) were employed. The scan time was about 19 min 44 s = 1.6 s (TR) × 2 (temporal interleave) × 2 (spatial interleave) × 2 (MEGA‐ON–OFF) × 16 (averages) × 12 (phase‐encoding steps)/half (z–k‐space weighting).
Figure 1.
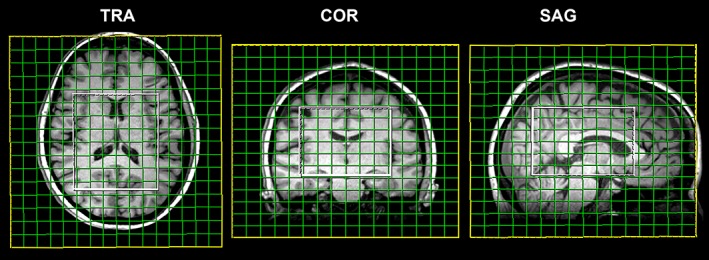
Morphological T 1‐weighted reference images display the positioning of the VOI (80 × 85 × 60 mm3) and FOV (200 × 200 × 170 mm3) in the transversal, coronal, and sagittal planes. The acquired matrix size was 14 × 14 × 12, with a nominal voxel size of about 14.3 × 14.3 × 14.3 mm3
Figure 2.
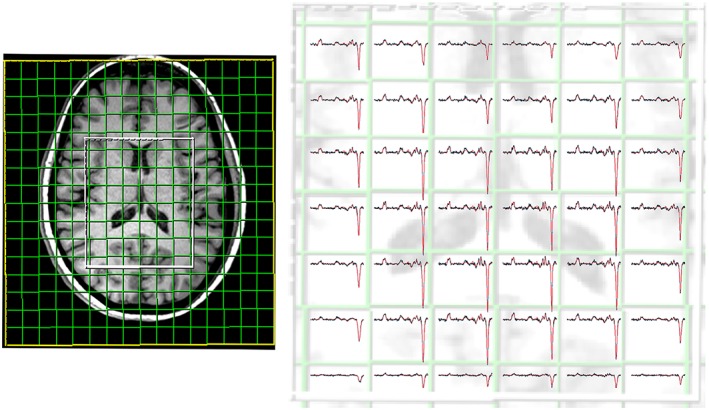
Morphological T 1‐weighted reference image display matrix and VOI (80 × 85 × 60 mm3) position in the transversal plane. The acquired matrix size was 14 × 14 × 12, with a nominal voxel size of about 14.3 × 14.3 × 14.3 mm3. On the right is a representative spectral grid displayed for the difference spectral dataset of GABA‐edited MEGA‐LASER 3D‐MRSI. All spectra in grids are scaled to the same noise level
2.4. Data processing
All spectra within the VOI were processed automatically with an in‐house‐developed software tool using MATLAB (R2013a, MathWorks, Natick, MA, USA), Bash (version 4.2.25, Free Software Foundation, Boston, MA, USA), and MINC (MINC Tools, Version 2.0; McConnell Brain Imaging Center, Montreal, QC, Canada), which features a graphical user interface for automatic data processing and employs LCModel software (Version 6.3–1, S. Provencher, LCModel, Oakville, ON, Canada). Head movement and B0‐changes were recorded for all volunteers and the average change in translation, rotation, frequency, and first‐order shim was calculated. All basis‐set simulations were performed with exactly the same RF and gradient modulations as in the experiments. Slice selection and gradient effects were simulated using a number of isochromats equal to the number of points in the gradient waveform (i.e. 350 points for the 3.5 ms GOIA pulse, 10 μs dwell time). Two different simulated basis sets without MM baseline correction were created using GAMMA, one for the EDIT‐OFF (containing 21 brain metabolites) and one for the difference spectrum (containing GABA+, Glx, and NAA) (Figure 3).21 The metabolic ratios (i.e. GABA+/Glx, GABA+/tCr, GABA+/tNAA, Glx/tCr, and Glx/tNAA), as well as spectral quality parameters (i.e. signal‐to‐noise ratio (SNR) and full width at half maximum (FWHM) of tNAA), were calculated. Maps of the metabolites' signal amplitudes (Figure 4) and their CRLBs (Cramér–Rao lower bounds) were created, along with FWHM and SNR maps of tNAA (Figure 5). For visualization, all metabolite maps were interpolated to the resolution of the MPRAGE images.
Figure 3.
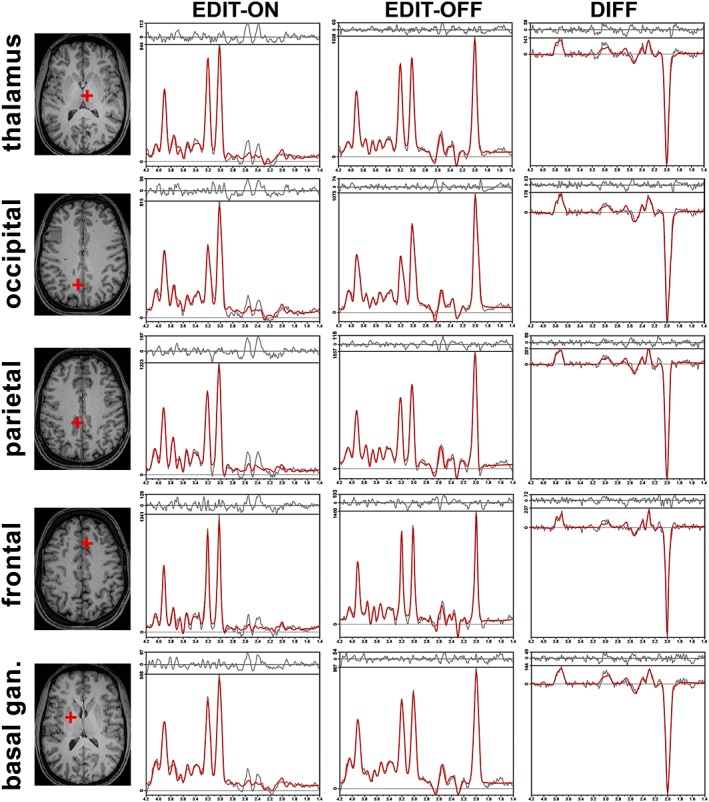
Samples of in vivo proton MR spectra obtained with the GABA‐editing MEGA‐LASER 3D MRSI sequence. In the first column, localizations of the selected voxel on T 1‐weighted images are shown in the transversal plane; in the second, third, and fourth columns, the LCModel fit of metabolites is shown in the EDIT‐ON (frequency suppression at 1.9 ppm), EDIT‐OFF (frequency suppression at 7.5 ppm), and DIFF (difference spectrum; subtraction of EDIT‐ON and EDIT‐OFF) spectrum, respectively
Figure 4.
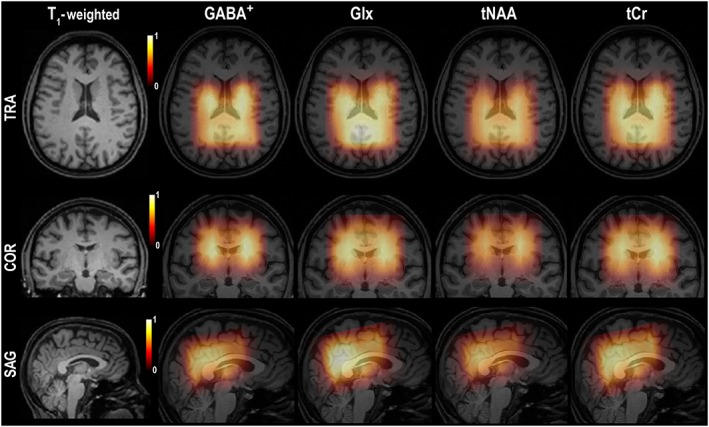
The first column displays the positioning of the VOI/FOV using T 1‐weighted images in all three orthogonal planes. Other columns (left–right) show 3D metabolic maps of GABA+ and Glx from the difference spectral dataset, as well as tNAA and tCr from the EDIT‐OFF spectral dataset. These were obtained from measurements in a volunteer, with GABA‐edited MEGA‐LASER, 3D MRSI. For visualization, all metabolite maps were interpolated to the resolution of the MPRAGE images
Figure 5.
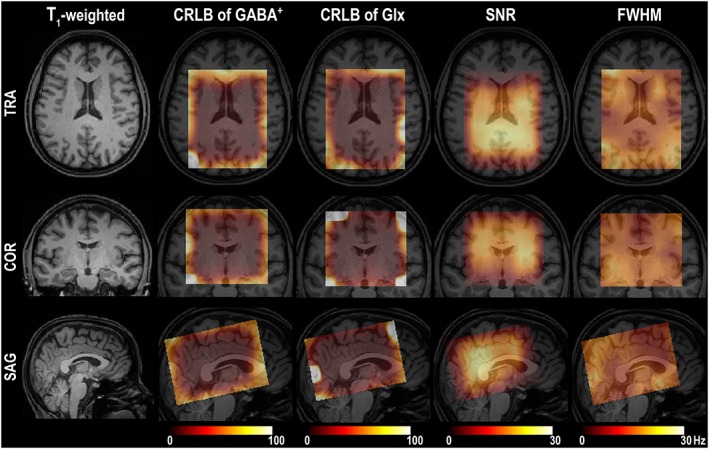
Images in all three orthogonal planes show 3D quality maps of a GABA‐edited, MEGA‐LASER, 3D MRSI sequence. The first column displays the positioning of the VOI/FOV using T 1‐weighted MRI. Other columns (left–right) illustrate 3D‐CRLB maps of GABA+ and Glx, SNR, and FWHM of tNAA, interpolated to the resolution of the MPRAGE images
2.5. Statistical evaluation
Statistical analysis was performed and plots were created using the SPSS software package (Version 15.0; Chicago, IL, USA). For quantitative evaluation, voxels inside the VOI that fulfilled the following minimum criteria were further processed: the CRLB of tNAA and the tCr in EDIT‐OFF spectra were less than 20% and the CRLB of GABA+ in the difference spectra was less than 30%. In all tests, descriptive parameters (i.e. mean; standard deviation (SD); coefficient of variation (CV)) of GABA+ and Glx metabolic ratios, as well as the SNR and FWHM of tNAA, were investigated. P values <0.05 were considered statistically significant. The intra‐subject reliability of GABA+ and Glx ratios between the test–retest sessions were evaluated across all eligible voxels for each volunteer by computing intraclass correlation coefficients (ICCs) using a two‐factor, mixed‐effects model and type consistency. Furthermore, to investigate the agreement between repeated 3D‐MRSI measurements of neurotransmitters, Bland–Altman plots were drawn for GABA+ and Glx ratios for all eligible voxels and volunteers (Supplementary Material S1). To analyze the difference in neurotransmitters between selected brain regions, one‐way ANOVA, followed by Bonferroni's post‐hoc tests, was performed across all volunteers. ANOVA was used to investigate differences between contralateral regions, gender, and test–retest sessions in neurotransmitter ratios, for each investigated brain region in all study participants.
3. RESULTS
Across all volunteer measurements, during the MEGA‐LASER 3D‐MRSI scan, a translation of 1.9 ± 0.8 mm, a rotation of 1.2 ± 0.7°, a frequency increase of 13.5 ± 2.2 Hz, and a B 0‐shim change of 3.5 ± 1.5 Hz/cm were recorded.
From a total of 216 voxels per VOI (in one case 288), 92 ± 7% (range 82–100%) of the voxels fulfilled the spectral quality criteria. The average SNR of tNAA in the EDIT‐OFF spectra was 16.6 ± 1.4 (mean ± SD), with an average FWHM for tNAA of 8.6 ± 0.8 Hz (mean ± SD).
The test–retest intra‐subject variability of GABA+ and Glx ratios, as well as CV and ICC, were evaluated for each volunteer across all eligible voxels (178–240 voxels) and are provided in Table 1. Low CVs and high ICCs for GABA+ ratios (CV, mean of medians 8%–ICC, mean > 0.75), as well as for Glx ratios (CV, mean of medians 6%–ICC, mean > 0.70) were found. Bland–Altman plots of test–retest reproducibility investigations are displayed in Supplementary Material S1. Inter‐subject values for CV in all 2835 eligible voxels over the whole group of subjects (i.e. 178–240 eligible voxels per VOI and per volunteer) showed low median values of about 8% for GABA+ ratios, as well as about 6% for Glx ratios (Table 2).
Table 1.
CV and ICC values for intra‐subject test–retest variability of GABA+ and Glx ratios evaluated for each volunteer across all eligible voxels (in each volunteer n = 178–240 voxels). Minimal (MIN), maximal (MAX), and averaged (mean with SD) values of ICC are shown, as well as medians of CV values calculated for each volunteer
| 14 volunteers (178–240 voxels) | CV (%) | ICC | ||||||
|---|---|---|---|---|---|---|---|---|
| MIN median | MAX median | Mean of medians | SD of medians | MIN | MAX | Mean | SD | |
| GABA + /Glx | 7.3 | 10.9 | 8.8 | 1.1 | 0.713 | 0.901 | 0.813 | 0.048 |
| GABA + /tCr | 6.3 | 9.8 | 8.0 | 1.2 | 0.674 | 0.863 | 0.795 | 0.049 |
| GABA + /tNAA | 5.9 | 9.5 | 7.5 | 1.0 | 0.661 | 0.841 | 0.764 | 0.050 |
| Glx/tCr | 4.0 | 7.8 | 5.8 | 1.1 | 0.569 | 0.846 | 0.703 | 0.091 |
| Glx/tNAA | 4.1 | 9.2 | 6.0 | 1.3 | 0.716 | 0.954 | 0.857 | 0.067 |
Table 2.
CV values for inter‐subject test–retest variability of GABA+ and Glx ratios evaluated for the whole group of volunteers across all eligible voxels (n = 2835 voxels in test, as well as in retest). Medians are shown with 25% and 75% of CV values
| CV (%) | |||
|---|---|---|---|
| Median | 25% percentile | 75% percentile | |
| GABA + /Glx | 8.7 | 4.1 | 15.7 |
| GABA + /tCr | 8.1 | 3.6 | 14.1 |
| GABA + /tNAA | 7.4 | 3.4 | 12.9 |
| Glx/tCr | 5.6 | 2.5 | 10.0 |
| Glx/tNAA | 5.9 | 2.6 | 10.7 |
There were 56 (14 (volunteers) × 2 (right/left) × 2 (test/retest)) voxels investigated for each of the following regions: the thalamus, the basal ganglia (i.e. globus pallidus), the frontal (i.e. anterior cingulated gyrus; Brodmann Area 24), the parietal (i.e. posterior cingulated gyrus; Brodmann Area 23), and the occipital lobe (i.e. parieto‐occipital sulcus; border of the medial part of the occipital lobe and the precuneus) (Figure 3). For each region, neurotransmitter ratios, descriptive statistics, and visualization of intra−/inter‐subject regional variability, as well as test–retest differences in selected brain regions, are shown in Figure 6. Figure 6 shows the low spatial variations of metabolite ratios across the brain (i.e. no significantly different levels in the contralateral brain regions), and the high reproducibility of repeated measurements (i.e. no significant differences in test–retest values). Table 3 lists the p values for differences between selected brain regions. Similar values were found for the GABA+/Glx ratios in the basal ganglia and thalamus, as well as in the parietal, frontal, and occipital regions, but all three cortical regions had significantly lower ratios than those in the thalamus and the basal ganglia. Likewise, the GABA+/tNAA ratio was significantly higher in the basal ganglia than in the thalamus, and in both areas it was higher than that in the remaining brain regions. No difference in the GABA+/tNAA ratio was found between the cortical regions. The frontal cortex had significantly lower levels of GABA+/tCr than all the other analyzed regions, with no significant differences among those regions. Similar levels of Glx/tNAA were detected in the frontal and occipital regions and both were significantly higher than those in the thalamus, the basal ganglia, and the parietal region. The thalamus, the basal ganglia, and the parietal region showed similar Glx/NAA ratios. The occipital region reached the highest and the basal ganglia the lowest levels of the Glx/tCr ratios among the selected brain areas, whereas no significant differences were found between the thalamus, the frontal region, or the parietal region. Furthermore, no significant gender‐related differences were found in neurotransmitter ratios across the selected brain regions (Supplementary Material S2).
Figure 6.
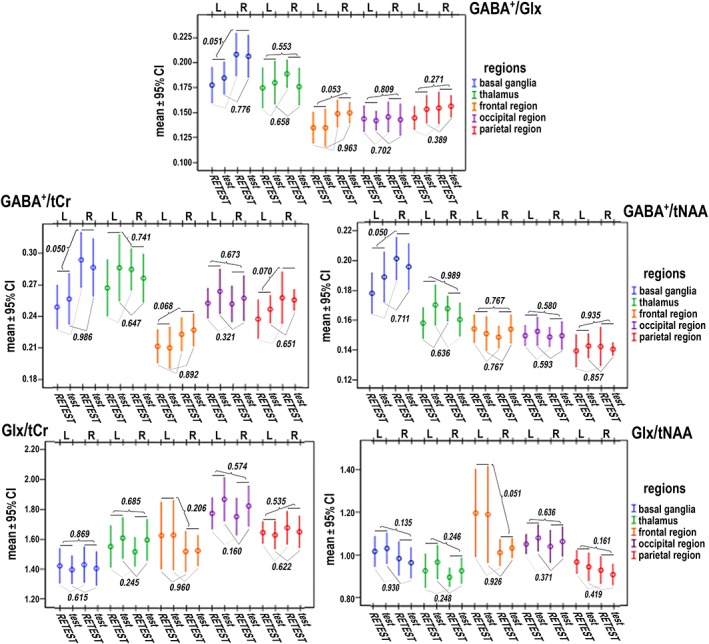
Graphs for intra−/inter‐subject variability of GABA+ and Glx ratios (mean ± 95% confidence interval) evaluated in selected brain regions (n = 56 in each region) with regard to spatial distribution (contralateral regions: 28 voxels for R (right), 28 voxels for L (left)) and repetition of measurement (28 voxels for test, 28 voxels for retest). P values obtained by ANOVA for differences in neurotransmitter ratios between contralateral brain regions (listed above the scatter bars) and test–retest sessions (listed under the scatter bars) are shown
Table 3.
p values expressing statistical differences in neurotransmitter ratios between selected brain regions (56 voxels in thalamus, basal ganglia, frontal, parietal, and occipital lobe) performed using one‐way ANOVA followed by Bonferroni's post‐hoc tests
| Ratios | p values | Basal ganglia | Frontal lobe | Parietal lobe | Occipital lobe |
|---|---|---|---|---|---|
| GABA + /Glx | thalamus | 0.070 | <0.001 | <0.001 | <0.001 |
| basal ganglia | <0.001 | <0.001 | <0.001 | ||
| frontal lobe | 0.549 | 1.000 | |||
| parietal lobe | 0.988 | ||||
| GABA + /tNAA | thalamus | <0.001 | 0.007 | <0.001 | 0.001 |
| basal ganglia | <0.001 | <0.001 | <0.001 | ||
| frontal lobe | 0.067 | 1.000 | |||
| parietal lobe | 0.128 | ||||
| GABA + /tCr | thalamus | 1.000 | <0.001 | 0.097 | 0.090 |
| basal ganglia | <0.001 | 0.097 | 1.000 | ||
| frontal lobe | <0.001 | <0.001 | |||
| parietal lobe | 1.000 | ||||
| Glx/tNAA | thalamus | 0.205 | <0.001 | 1.000 | <0.001 |
| basal ganglia | 0.004 | 0.417 | 0.048 | ||
| frontal lobe | <0.001 | 1.000 | |||
| parietal lobe | 0.001 | ||||
| Glx/tCr | thalamus | 0.003 | 1.000 | 0.539 | <0.001 |
| basal ganglia | 0.002 | <0.001 | <0.001 | ||
| frontal lobe | 0.725 | <0.001 | |||
| parietal lobe | 0.003 |
Test–retest variability in the GABA+ and Glx ratios for all brain regions are displayed in Table 4. The low median CV values of GABA+ (6–9%) and Glx (2–8%) ratios demonstrate the high spatial reproducibility of the measurements.
Table 4.
CV values (%) for test–retest variability of GABA+ and Glx ratios in selected brain regions across all volunteers (n = 56 voxels in each region). Medians are shown with 25% and 75% of CV values
| CV (56 voxels) | GABA + /Glx | GABA + /tCr | GABA + /tNAA | Glx/tCr | Glx/tNAA | |||||
|---|---|---|---|---|---|---|---|---|---|---|
| Median | 25–75% percentile | Median | 25–75% percentile | Median | 25–75% percentile | Median | 25–75% percentile | Median | 25–75% percentile | |
| Thalamus | 8 | 4–15 | 7 | 4–13 | 8 | 3–12 | 8 | 2–11 | 8 | 2–11 |
| Basal g. | 9 | 5–15 | 9 | 3–16 | 7 | 5–13 | 7 | 2–13 | 7 | 3–15 |
| Frontal | 9 | 4–15 | 6 | 3–12 | 6 | 2–8 | 4 | 2–6 | 5 | 2–10 |
| Parietal | 7 | 3–13 | 7 | 2–12 | 7 | 3–10 | 4 | 3–8 | 5 | 3–8 |
| Occipital | 6 | 3–9 | 6 | 4–9 | 8 | 4–10 | 2 | 2–5 | 3 | 2–5 |
4. DISCUSSION
This work evaluated the spatial distribution of neurotransmitter levels and intra−/inter‐subject variability for 3D mapping of GABA+ and Glx in vivo, using proton MRSI of the human brain at 3 T. This study provides evidence that the MEGA‐LASER‐edited 3D‐MRSI sequence enables highly stable results, which makes it suitable for clinical and neuroscientific studies.
4.1. Comparison with other sequence approaches
The GABA‐edited, MEGA‐LASER‐based, 3D‐MRSI sequence described here acquires voxels of about 3 cm3 nominal volume in about 20 min. To date, GABA‐edited MRS in the human brain has been performed mostly via single‐voxel spectroscopy (SVS) in about 8–17 min,4, 26, 27 and in a few cases via single‐slice 2D‐MRSI in about 17–35 min,5, 21 with voxel sizes in the range of 1–8 cm3.
GABA‐edited MRS is most commonly implemented using MEGA editing combined with point resolved spectroscopy (PRESS) localization, the so‐called MEGA‐PRESS.26, 28 The MEGA‐LASER approach used here provides higher SNR and better localization accuracy, as reported recently.20, 21
In addition, this study used spiral‐encoded 3D‐MRSI combined with efficient real‐time motion‐ and scanner drift‐related artifact correction, which enables fast and robust measurements.20, 23 High spectral quality was achieved consistently in each VOI in this study (i.e. ~200 voxels per subject), which guaranteed high intra‐subject repeatability. These results indicate that the described MEGA‐LASER 3D‐MRSI is a robust method for the in vivo detection of GABA+, as well as Glx.
Despite the increasingly widespread use of J‐difference‐edited MRS techniques, the accuracy and reproducibility of the GABA and simultaneously measured Glx signal has only rarely been demonstrated.17, 29, 30 In the present study, the intra‐ and inter‐subject variability of GABA+ and Glx ratios (i.e. GABA+/Glx, GABA+/tNAA, GABA+/tCr, Glx/tNAA, and Glx/tNAA) was investigated. Although the concentration of tNAA and tCr may vary in the human brain, the metabolite assessment using their ratios is considered clinically suitable and widely applicable.5, 31
4.2. Intra‐subject test–retest variability
The intra‐subject CV for repeated scans in each of the 14 volunteers was, on average, a median of about 8% for GABA+ ratios, and about 6% for Glx ratios, suggesting that the described GABA‐edited 3D‐MRSI measurements were highly reproducible. Glx measurements were similarly repeatable, with Glx CVs of 4–8% (minimal–maximal value of medians, for Glx/tCr), and 4–9% (for Glx/tNAA), whereas GABA+ ratios were more variable, ranging from 6 to 10% (both GABA+/tNAA and GABA+/tCr) and 7–11% CVs for GABA+/Glx. These values are consistent with those reported for other GABA J‐difference‐editing MRS studies (i.e. usually SVS, MEGA‐PRESS), which reported intra‐individual variability of repeated GABA+ measurements in the range of 7–15%,14, 27, 30 whereas Glx was in the range of 6–18%.1, 17, 32 A high degree of reliability between test–retest measurements was also confirmed using ICC analysis.
4.3. Inter‐subject variability
The inter‐subject variability of GABA+ and Glx ratios (median CV for GABA+ ratios about 8%; for Glx ratio about 6%) described here were similar to the measured test–retest intra‐subject variability. This may indicate that the reproducibility of metabolite measurements is predominantly determined by MR scanner features (e.g. B 0 field inhomogeneities, scanner drift or heating, gradient coil instability) and by individual scanning factors (e.g. positioning, in/voluntary movement, tissue inhomogenities)27, 28 and not by concentration differences between healthy subjects. All these aspects have a significant impact on test–retest reproducibility assessment. Undesirable effects that would increase the intra‐subject CV were limited as much as possible by using auto‐align (i.e. automated repositioning of the MRSI volume with sub‐millimeter accuracy between scans), adiabatic refocusing pulses (i.e. elimination of B 1 + sensitivity and chemical‐shift displacement errors), and by the application of real‐time movement, shim, and frequency artifact correction.20, 21 In addition, an appropriate coil combination, based on the work of Roemer et al., was employed.22 The remaining spatial signal variations were removed when calculating metabolite ratios.33 The automatically performed frequency drift correction prevented the editing pulses from slowly drifting away from the GABA resonance at 1.9 ppm.3 The neurotransmitter reproducibility achieved in this study is comparable to or higher than that reported in other experimental studies. The measured test–retest inter‐subject variability of GABA+ ratios in this study (i.e. CV for GABA+/Glx ~ 9%, for GABA+/tCr ~ 8%, for GABA+/tNAA ~7%) is slightly lower than the values of 10–15% published in other studies based on SVS and MEGA‐PRESS.14, 29, 32 The Glx reproducibility values derived here with MEGA‐LASER 3D‐MRSI (~6% CVs for Glx ratios) are consistent with those reported using MEGA‐PRESS 1H MRS (CVs 5–16%).17, 32, 34 A Bland–Altman analysis supported the high agreement between repeated 3D‐MRSI measurements of GABA+ and Glx ratios.
4.4. Spatial and gender‐related neurotransmitter variations
This study confirms regional differences in neurotransmitter concentrations within the CNS, as was previously suggested.2, 5, 35 The GABA+ ratios reported here were higher in the basal ganglia and thalamus than in the parietal, frontal, and occipital regions. However, the spectral quality was somewhat lower in the frontal lobe, which could explain the larger variability in this region. Only GABA+/tCr ratios showed significantly reduced values in the frontal cortex relative to the occipital and parietal regions, possibly because tCr is higher in the frontal areas.2, 31 Likewise other studies reported higher GABA+ ratios in the basal ganglia compared with the frontal, occipital, or temporal regions,26 but lower GABA+ ratios in the frontal compared with the occipital region,26, 36 that were still higher than in the parietal region.4 This may reflect differences in the white matter (WM) and gray matter (GM) composition of the selected brain tissue. In this study higher Glx/tNAA levels in occipital and frontal regions compared with the parietal cortex, the basal ganglia, and the thalamus were also detected. Among all selected brain regions the highest Glx/tCr ratios were found in the occipital region and the lowest in the basal ganglia, in agreement with previous reports.17, 35 Higher Glx levels in the frontal compared with the parietal region are also consistent with previous observations.4, 35 Other authors have found high Glx in cortical areas,17, 37 but this may be attributable to GM/WM‐related differences in tCr.35, 37
Although the CVs in various regions were very similar, the assessment of neurotransmitter ratios in the thalamus and basal ganglia (Glx ratios 7–8%; GABA+ ratios 7–9%) were slightly less repeatable compared with the frontal, parietal, and occipital lobes (2%–5% for Glx ratios; 6%–8% for GABA+ ratios). These results indicate that the applied MEGA‐LASER 3D‐MRSI sequence provides reliable GABA+ and Glx measurements across the investigated brain regions, even in the presence of varying spectral quality, which is essential for clinical and neuroscientific studies.
In 1H MRS studies, gender‐related differences in neurotransmitter concentrations have been inconsistent, demonstrating no, or only regional, changes in GABA or in Glx levels.17, 35, 38 These discrepancies could be caused by hormonal differences between subjects.16, 17 In this study there were no gender differences observed in neurotransmitter ratios. The variability of neurotransmitter ratios in the male and female subgroups was similar (i.e., the differences in CV did not exceed 3%).
4.5. Limitations
The methodology of this study has limitations. First, the LASER‐selected VOI in this study was focused on a relatively small central brain area. Another restriction is the use of metabolite ratios5, 31 rather than absolute quantification using internal water referencing.31 In future studies a matching water reference scan could be obtained in 1 min 23 s when reducing the averages from 16 to 1. Second, no correlation between tissue composition and neurotransmitter levels was investigated, but rather differences in the concentrations of GABA+, as well as Glx, between different brain regions. Third, the MM contamination of the quantified GABA+ signal leads to overestimation of GABA and its ratios, which should be corrected in future studies. However, the MM contribution is constant in all subjects over time,17 and measurement approaches that correct for MM contributions are challenging.39 For MEGA‐LASER or MEGA‐SPECIAL, MM contamination was reported in the range of 34–50% (References 39 and 40). For MEGA‐LASER, this assessment remains to be performed.
5. CONCLUSIONS
The results of this study show that spiral‐encoding, GABA‐edited, MEGA‐LASER 3D‐MRSI can be a fast, robust, and reproducible method for in vivo GABA+ and Glx mapping on high‐field (3 T) MR scanners. This method has the potential to be used in neuroscientific and clinical studies to detect pathological spatial alterations of GABA+ and Glx in the brain.
Supporting information
Supplementary Material
Supporting information
ACKNOWLEDGMENTS
This study was supported by the Austrian Science Fund (FWF), KLI‐61, and the FFG Bridge Early Stage Grant 846505, and by the projects SAS–NSC Joint Research Cooperation Grant 2013/17, VEGA 2/0191/15, “Increasing opportunities for career growth in research and development in the field of medical sciences” (ITMS: 26110230067), and “Biomedical Center Martin” (ITMS: 26220220187), co‐funded from EU sources.
Hnilicová, P. , Považan, M. , Strasser, B. , Andronesi, O. C. , Gajdošík, M. , Dydak, U. , Ukropec, J. , Dobrota, D. , Trattnig, S. , and Bogner, W. (2016) Spatial variability and reproducibility of GABA‐edited MEGA‐LASER 3D‐MRSI in the brain at 3 T. NMR Biomed., 29: 1656–1665. doi: 10.1002/nbm.3613.
REFERENCES
- 1. Agarwal N, Renshaw PF. Proton MR spectroscopy‐detectable major neurotransmitters of the brain: biology and possible clinical applications. Am J Neuroradiol. 2012;33:595–602. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Haga KK, Khor YP, Farrall A, Wardlaw JM. A systematic review of brain metabolite changes, measured with 1H magnetic resonance spectroscopy, in healthy aging. Neurobiol Aging. 2009;30:353–363. [DOI] [PubMed] [Google Scholar]
- 3. de Graaf RA. In Vivo NMR Spectroscopy. Chichester, UK: Wiley;2007. [Google Scholar]
- 4. Gao F, Edden RA, Li M, et al. Edited magnetic resonance spectroscopy detects an age‐related decline in brain GABA levels. Neuroimage. 2013;78:75–82. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Puts NA, Edden RA. In vivo magnetic resonance spectroscopy of GABA: a methodological review. Prog Nucl Magn Reson Spectrosc. 2012;60:29–41. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Lanctot KL, Herrmann N, Mazzotta P, Khan LR, Ingber N. GABAergic function in Alzheimer's disease: evidence for dysfunction and potential as a therapeutic target for the treatment of behavioural and psychological symptoms of dementia. Can J Psychiatry. 2004;49:439–453. [DOI] [PubMed] [Google Scholar]
- 7. Pfeiffer R, Wszolek Z, Ebadi M. Parkinson's Disease. 3rd ed. Boca Raton, Florida, USA: CRC Press, Taylor & Francis Group;2012. [Google Scholar]
- 8. Stagg CJ, Lang B, Best JG, et al. Autoantibodies to glutamic acid decarboxylase in patients with epilepsy are associated with low cortical GABA levels. Epilepsia. 2010;51:1898–1901. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Pollack MH, Jensen JE, Simon NM, Kaufman RE, Renshaw PF. High‐field MRS study of GABA, glutamate and glutamine in social anxiety disorder: response to treatment with levetiracetam. Prog Neuropsychopharmacol Biol Psychiatry. 2008;32:739–743. [DOI] [PubMed] [Google Scholar]
- 10. Levy LM, Degnan AJ. GABA‐based evaluation of neurologic conditions: MR spectroscopy. Am J Neuroradiol. 2013;34:259–265. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Chang L, Cloak CC, Ernst T. Magnetic resonance spectroscopy studies of GABA in neuropsychiatric disorders. J Clin Psychiatry. 2003;64:7–14. [PubMed] [Google Scholar]
- 12. Tayoshi S, Nakataki M, Sumitani S, et al. GABA concentration in schizophrenia patients and the effects of antipsychotic medication: a proton magnetic resonance spectroscopy study. Schizophr Res. 2010;117:83–91. [DOI] [PubMed] [Google Scholar]
- 13. Rowland LM, Kontson K, West J, et al. In vivo measurements of glutamate, GABA, and NAAG in schizophrenia. Schizophr Bull. 2013;39:1096–1104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Bogner W, Gruber S, Doelken M, et al. In vivo quantification of intracerebral GABA by single‐voxel 1H‐MRS—how reproducible are the results? Eur J Radiol. 2010;73:526–531. [DOI] [PubMed] [Google Scholar]
- 15. Novotny EJ Jr, Fulbright RK, Pearl PL, Gibson KM, Rothman DL. Magnetic resonance spectroscopy of neurotransmitters in human brain. Ann Neurol. 2003;54:S25–S31. [DOI] [PubMed] [Google Scholar]
- 16. Cleve M, Gussew A, Reichenbach JR. In vivo detection of acute pain‐induced changes of GABA+ and Glx in the human brain by using functional 1H MEGA‐PRESS MR spectroscopy. Neuroimage. 2015;105:67–75. [DOI] [PubMed] [Google Scholar]
- 17. O'Gorman RL, Michels L, Edden RA, Murdoch JB, Martin E. In vivo detection of GABA and glutamate with MEGA‐PRESS: reproducibility and gender effects. J Magn Reson Imaging. 2011;33:1262–1267. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Mullins PG, McGonigle DJ, O'Gorman RL, et al. Current practice in the use of MEGA‐PRESS spectroscopy for the detection of GABA. Neuroimage. 2014;86:43–52. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Evans CJ, McGonigle DJ, Edden RA. Diurnal stability of gamma‐aminobutyric acid concentration in visual and sensorimotor cortex. J Magn Reson Imaging. 2010;31:204–209. [DOI] [PubMed] [Google Scholar]
- 20. Bogner W, Hess AT, Gagoski B, et al. Real‐time motion‐ and B0‐correction for LASER‐localized spiral‐accelerated 3D‐MRSI of the brain at 3 T. Neuroimage. 2014;88:22–31. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Bogner W, Gagoski B, Hess AT, et al. 3D GABA imaging with real‐time motion correction, shim update and reacquisition of adiabatic spiral MRSI. Neuroimage. 2014;103:290–302. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Roemer PB, Edelstein WA, Hayes CE, Souza SP, Mueller OM. The NMR phased array. Magn Reson Med. 1990;16:192–225. [DOI] [PubMed] [Google Scholar]
- 23. Andronesi OC, Gagoski BA, Sorensen AG. Neurologic 3D MR spectroscopic imaging with low‐power adiabatic pulses and fast spiral acquisition. Radiology. 2012;262:647–661. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Andronesi OC, Ramadan S, Ratai EM, Jennings D, Mountford CE, Sorensen AG. Spectroscopic imaging with improved gradient modulated constant adiabaticity pulses on high‐field clinical scanners. J Magn Reson. 2010;203:283–293. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. van der Kouwe AJ, Benner T, Fischl B, et al. On‐line automatic slice positioning for brain MR imaging. Neuroimage. 2005;27:222–230. [DOI] [PubMed] [Google Scholar]
- 26. Durst CR, Michael N, Tustison NJ, et al. Noninvasive evaluation of the regional variations of GABA using magnetic resonance spectroscopy at 3 Tesla. Magn Reson Imaging. 2015;33:611–617. [DOI] [PubMed] [Google Scholar]
- 27. Near J, Ho YC, Sandberg K, Kumaragamage C, Blicher JU. Long‐term reproducibility of GABA magnetic resonance spectroscopy. Neuroimage. 2014;99:191–196. [DOI] [PubMed] [Google Scholar]
- 28. Veenith TV, Mada M, Carter E, et al. Comparison of inter subject variability and reproducibility of whole brain proton spectroscopy. PLoS One. 2014;9: e115304 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Henry ME, Lauriat TL, Shanahan M, Renshaw PF, Jensen JE. Accuracy and stability of measuring GABA, glutamate, and glutamine by proton magnetic resonance spectroscopy: a phantom study at 4 Tesla. J Magn Reson. 2011;208:210–218. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Long Z, Dyke JP, Ma R, Huang CC, Louis ED, Dydak U. Reproducibility and effect of tissue composition on cerebellar gamma‐aminobutyric acid (GABA) MRS in an elderly population. NMR Biomed. 2015;28:1315–1323. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Barker PB, Bizzi A, Stefano ND, Gullapalli R, Lin DDM. Clinical MR Spectroscopy: Techniques and Applications. Cambridge: Cambridge University Press;2009. [Google Scholar]
- 32. Geramita M, van der Veen JW, Barnett AS, et al. Reproducibility of prefrontal gamma‐aminobutyric acid measurements with J‐edited spectroscopy. NMR Biomed. 2011;24:1089–1098. [DOI] [PubMed] [Google Scholar]
- 33. Gillard JH, Waldman AD, Barker PB. Clinical MR Neuroimaging: Physiological and Functional Techniques. Cambridge: Cambridge University Press;2010. [Google Scholar]
- 34. Wijtenburg SA, Rowland LM, Edden RA, Barker PB. Reproducibility of brain spectroscopy at 7 T using conventional localization and spectral editing techniques. J Magn Reson Imaging. 2013;38:460–467. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Sailasuta N, Ernst T, Chang L. Regional variations and the effects of age and gender on glutamate concentrations in the human brain. Magn Reson Imaging. 2008;26:667–675. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. van der Veen JW, Shen J. Regional difference in GABA levels between medial prefrontal and occipital cortices. J Magn Reson Imaging. 2013;38:745–750. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Kaiser LG, Schuff N, Cashdollar N, Weiner MW. Age‐related glutamate and glutamine concentration changes in normal human brain: 1H MR spectroscopy study at 4 T. Neurobiol Aging. 2005;26:665–672. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Hu Y, Chen X, Gu H, Yang Y. Resting‐state glutamate and GABA concentrations predict task‐induced deactivation in the default mode network. J Neurosci. 2013;33:18566–18573. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Near J, Simpson R, Cowen P, Jezzard P. Efficient gamma‐aminobutyric acid editing at 3 T without macromolecule contamination: MEGA‐SPECIAL. NMR Biomed. 2011;24:1277–1285. [DOI] [PubMed] [Google Scholar]
- 40. Waddell KW, Avison MJ, Joers JM, Gore JC. A practical guide to robust detection of GABA in human brain by J‐difference spectroscopy at 3 T using a standard volume coil. Magn Reson Imaging. 2007;25:1032–1038. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplementary Material
Supporting information


