Figure 3.
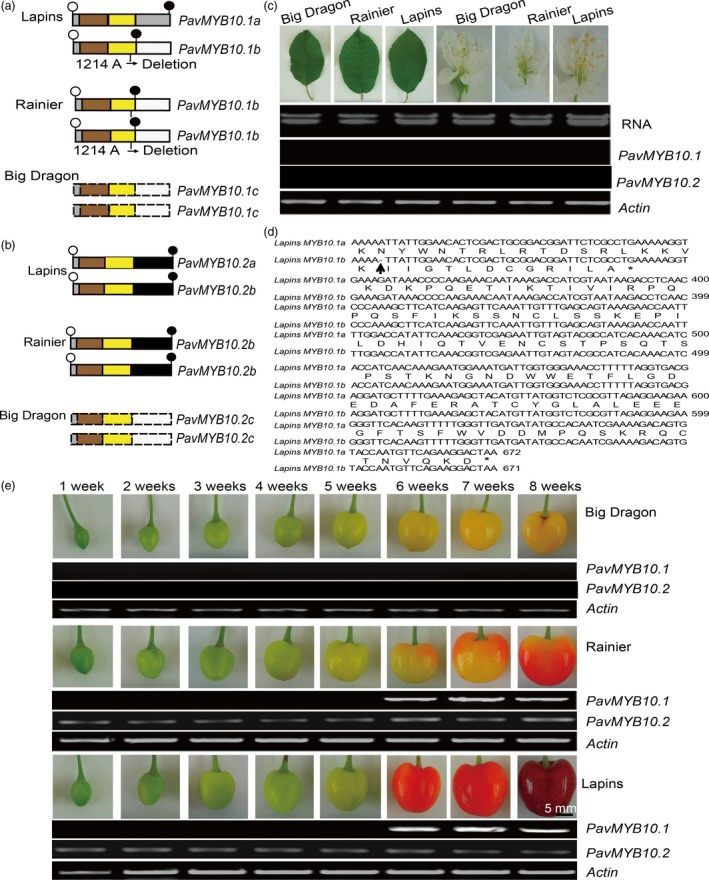
Transcript analysis of PavMYB10.1 and PavMYB10.2 in sweet cherry varieties. (a and b) Schematic of PavMYB10.1 and PavMYB10.2 transcript structure in ‘Lapins’, ‘Rainier’ and ‘Big Dragon’. Truncated PavMYB10.1b transcript was detectable in blush variety ‘Rainier’. PavMYB10.1a and PavMYB10.1b transcripts were detected in dark‐red variety ‘Lapins’. PavMYB10.2a and PavMYB10.2b were identical in ‘Lapins’ and ‘Rainier’. Dashed boxes indicate undetectable PavMYB10.1 and PavMYB10.2. Open and filled circles represent start and stop codons. Sienna and yellow boxes represent R2 and R3 domains. (c) Difference of the PavMYB10.1a and PavMYB10.1b sequence. Upwards arrow indicates position of deletion, asterisk marks stop codon. (d) RT‐PCR analysis of PavMYB10.1 and PavMYB10.2 transcript levels in leaves and flowers of ‘Big Dragon’, ‘Rainier’ and ‘Lapins’. Actin was used as loading control. (e) RT‐PCR analysis of PavMYB10.1 and PavMYB10.2 transcript levels during fruit development of ‘Big Dragon’, ‘Rainier’ and ‘Lapins’. Transcript levels of PavMYB10.1 and PavMYB10.2 from 1 to 8 weeks after full bloom in ‘Big Dragon’, ‘Rainier’ and ‘Lapins’ are shown. Actin was used as a loading control.
