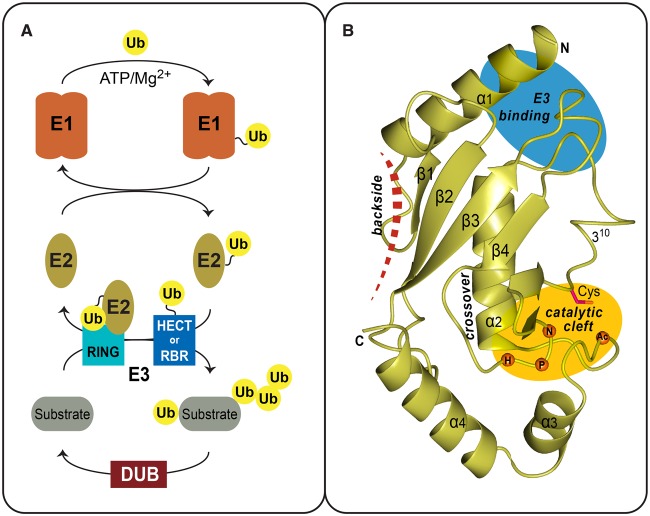
Figure 1. Ubiquitin pathway and the E2 fold.
(A) Overview of the Ub pathway and the enzymes involved at each step: activation (E1), conjugation (E2), ligation (E3), and deubiquitination (DUB). The E1 mediates ubiquitin activation in an energy-consuming step. The ubiquitin thioester is then transferred onto a catalytic cysteine of the E2 enzyme. RING-type E3s form a non-covalent complex with the E2∼Ub thioester intermediate or, alternatively, ubiquitin is transferred to catalytic sites of HECT and RBR-type E3 ligases. The E3 enzymes ultimately catalyze ubiquitination of a substrate lysine. Ubiquitin signals can also be extended to form polyubiquitin chains. Finally, DUBs catalyze the removal of ubiquitin. (B) Ribbon diagram of UBE2D2 (PDB 2ESK) as a representative of the UBC fold conserved among ubiquitin E2s. Secondary structure elements and the termini of the domain are indicated. Also depicted is the location of the active-site cysteine within the catalytic cleft (yellow oval), the E3-binding region (blue oval), the cross-over helix that stabilizes ‘closed’ conformers of the E2∼Ub thioester intermediate and location of the E2 backside-binding surface (red dashed line).