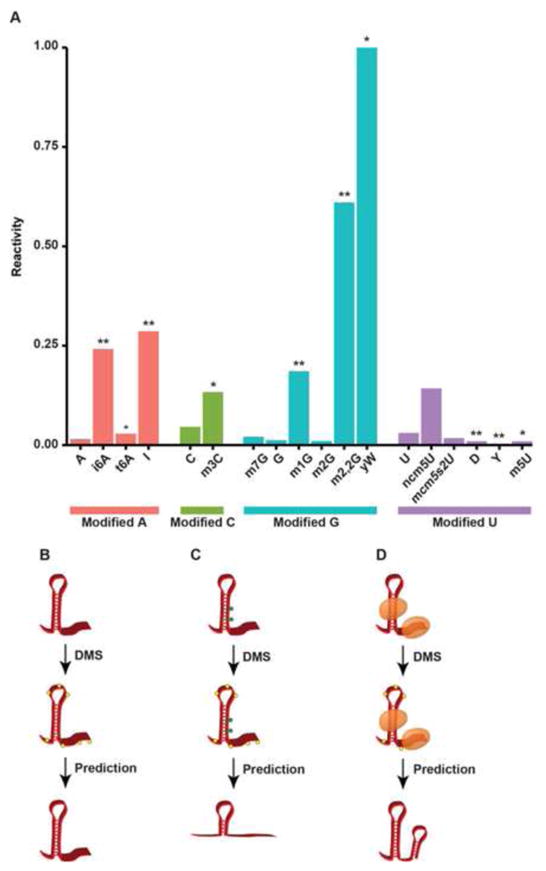
Figure 2. Covalent RNA modifications and limitations to Structure-seq.
(A) The stopping power for each covalently modified nucleotide, defined as the percentage of reads terminating via reverse transcriptase stalling normalized to total read coveraqe. * p < 0.05, ** p < 1×10−10. (B) An example of the ideal case for Structure-seq in which a transcript is unbound and has no covalently modified nucleotides. Yellow hexagons represent the DMS adduct in all figures. (C) An example of covalent modifications leading to increased RT stops, resulting in a predicted ssRNA sequence in a paired region. Green hexagons represent covalent modifcations to specific nucleotides. (D) A schematic showing proteins occluding DMS addition, leading to a predicted stretch of dsRNA in what is actually a single-stranded region.