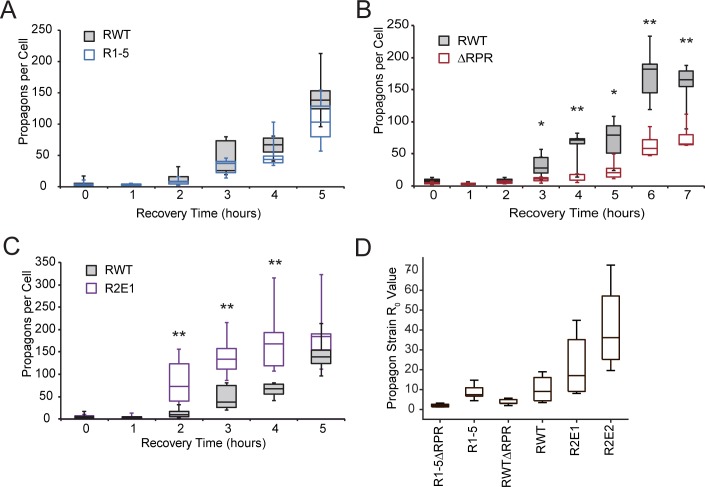
Fig 4. Sequence variant strains differ in their ability to amplify Sup35 amyloid.
A. Wildtype [PSI+] (SLL2606) and R1-5 (SY2022) strains were grown in YPAD + 3mM GdnHCl until nearly cured, followed by growth in YPAD for the indicated times. At each time point, the number of propagons per cell was determined. Horizontal lines on boxes indicate 25th, 50th, and 75th percentiles, and whiskers indicate maximum and minimum. n≥4. B. Wildtype [PSI+] (SLL2606) and ΔRPR (SY2023) strains were treated as in A. n≥4, *p<0.05, **p<0.01, student’s t-test. C. Wildtype [PSI+] (SLL2606) and R2E1 (SY2247) strains treated as in A. n≥4, **p<0.01, students t-test. D. Mathematical modeling was used to calculate the R0 value for the indicated strains using the steady-state soluble Sup35 levels (Fig 3B). Box plots are as described in panel A. Statistical analysis is described in the Supplementary Materials.