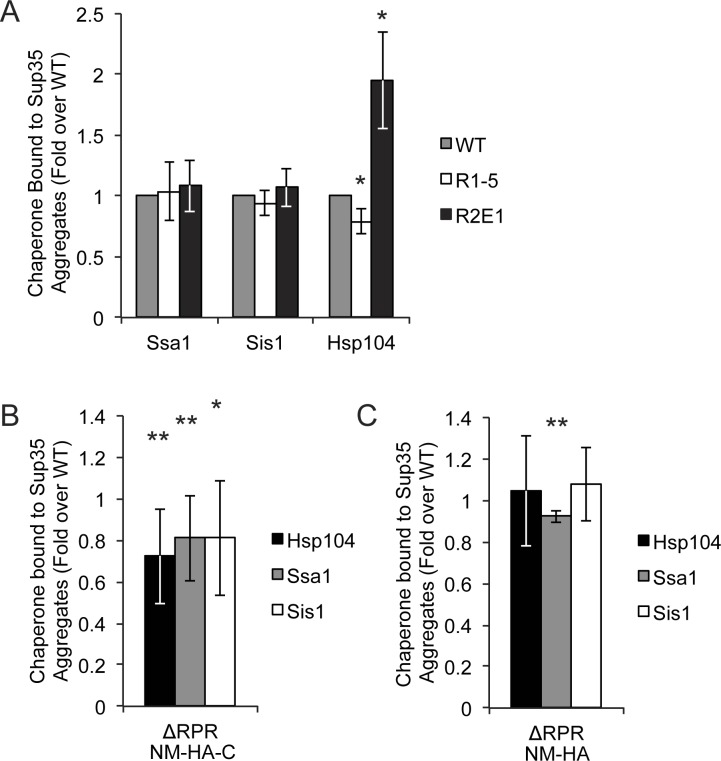
Fig 7. Chaperone binding to Sup35 amyloid differs in sequence variant strains.
A. NM-HA and co-aggregating full-length (untagged) Sup35 in wildtype (SY3007), R1-5 (SY3008), and R2E1 (SY3010), strains were immunocaptured using anti-HA magnetic beads and separated by SDS-PAGE, and the amount of bound chaperone proteins was quantified by immunoblotting and corrected for the amount of Sup35 present in aggregates in each strain (see methods). Bars represent means; error bars represent standard deviations, n≥4, *p<0.05, student’s t-test. B. NM-HA-C and co-aggregating full-length Sup35 in wildtype (SY3159) and ΔRPR (SY3164) strains were immunocaptured with anti-HA magnetic beads and separated by SDS-PAGE, and the amount of bound chaperones was quantified by immunoblotting and corrected for the amount of Sup35 present in aggregates. Bars represent means; error bars represent standard deviations. n≥8, *p<0.05, **p<0.01, student’s t-test. C. NM-HA and co-aggregating full-length (untagged) Sup35 in wildtype (SY3007) and ΔRPR (SY3009) strains were immunocaptured with anti-HA magnetic beads and separated by SDS-PAGE, and the amount of bound chaperones was quantified by western blot and corrected for the amount of Sup35 present in aggregates. Bars represent means; error bars represent standard deviations. n = 4, **p<0.01, student’s t-test.