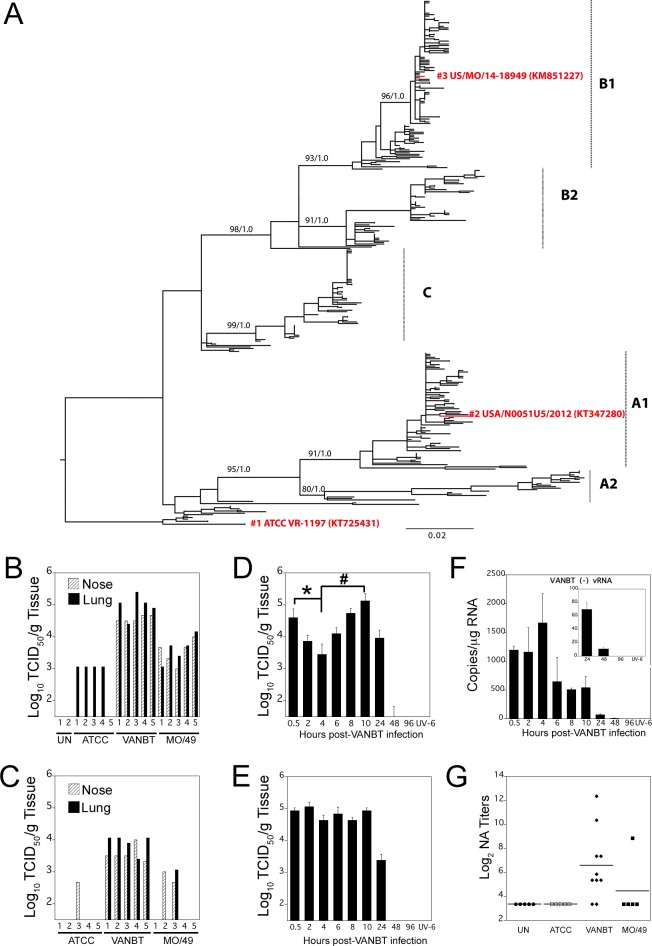
Fig 1. A cotton rat model of EV-D68 infection and replication.
Cotton rats were infected i.n. with 106 TCID50 per rat with 3 different strains of EV-D68, ATCC (prototype Fermon strain), VANBT (2012 pre-outbreak strain from Nashville, TN) and MO/49 (2014 outbreak strain from Kansas, MO). (A) Evolutionary tree showing three major clades of EV-D68, A, B and C, distributed worldwide. Strains used in this study are shown in red. The tree is rooted by the oldest EV-D68 sequence in GenBank, the Fermon strain (referred to as ATCC), collected in 1962 in California, USA. (B, C) Quantification of infectious virus titers of each EV-D68 strain in nose and lung homogenates from infected animals at 10 h (B) and 24 h (C) p.i. Groups of 5 animals were euthanized at each time point. Each bar corresponds to an individual animal. UN = uninfected. (D, E, F) Time course of VANBT replication in cotton rats; groups of 4 animals were euthanized at the indicated time to measure infectious virus titers in nose (D) and lung (E) homogenates. Results are representative of 2 independent experiments. *p<0.05 for 4 h nose titer compared with 0.5 h p.i. and #p<0.05 for 10 h nose titer compared with 4 h p.i. Animals inoculated with UV-VANBT and sacrificed at 6 h p.i. (UV-6) were shown as control. (F) Quantification of VANBT (-) vRNA by qRT-PCR in lung tissue. Insert is a blowup of the 24, 48 and 96 h time points from VANBT-infected and UV-VANBT-inoculated rat sacrificed at 6 h p.i. (G) Homologous NA titers measured in serum samples collected 3 weeks p.i. with the indicated EV-D68.