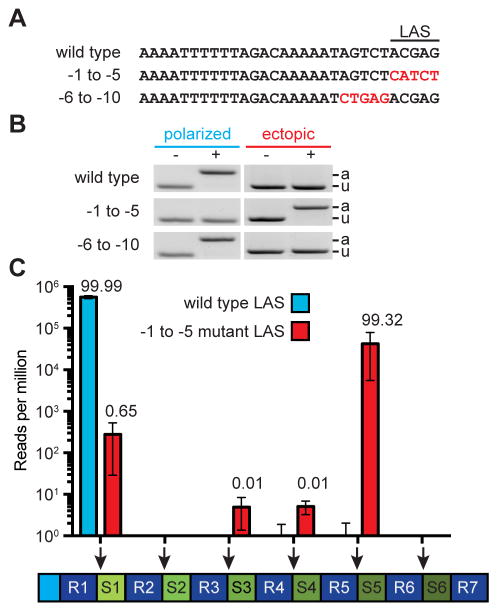
Figure 2. A sequence within leader specifies the site of spacer integration.
See also Fig. S1.
(A) Mutations were introduced at the 3′ end of the leader to define the leader-anchoring sequence (LAS).
(B) Strains containing the leader mutations described in (A) were infected with phage ϕNM4γ4 and analyzed by PCR for polarized and ectopic spacer integration.
(C) Analysis of the site of spacer integration using next-generation sequencing. Liquid cultures harboring a wild-type or mutant LAS were infected at a MOI of 1 the DNA isolated from surviving cells at the end of infection was used for PCR amplification of the array. The expanded PCR amplicons were purified from the gel and used for MiSeq next-generation sequencing. Bars shows the number of normalized reads for the integration of new spacers in each possible position of the CRIPSR array (marked by the red arrows). Mean ± S.E.M. of three replicas are reported.