Abstract
Surfactant protein B (SP-B; 79 residues) is a member of the saposin superfamily and plays a pivotal role in lung function. The N- and C-terminal regions of SP-B, cross-linked by two disulfides, were theoretically predicted to fold as charged amphipathic helices, suggesting participation in surfactant activities. Previous studies with oxidized Super Mini-B (SMB), a construct based on the N- and C-regions of SP-B (i.e., residues 1–25 and 63–78) joined with a designer turn (-PKGG-) and two disulfides, indicated that freshly prepared SMB in lipids folded as a surface active, α-helix-hairpin. Because other peptides modeled on α-helical SP domains lost helicity and surfactant activity on storage, experiments were here performed on oxidized SMB in surfactant liposomes stored at ~2–8°C for ≤5.5 years. Captive bubble surfactometry confirmed low minimum surface tensions for fresh and stored SMB preparations. FTIR spectroscopy of fresh and stored SMB formulations showed secondary structures compatible with the peptide folding as α-helix-hairpin. A homology (I-TASSER) model of oxidized SMB demonstrated a globular protein, exhibiting a core of hydrophobic residues and a surface of polar residues. Since mass spectroscopy indicated that the disulfides were maintained on storage, the stability of SMB may be partly due to the disulfides bringing the N- and C-α-helices closer. Mass spectroscopy of stored SMB preparations showed some methionine oxidation, and also partial deacylation of surfactant phospholipids to form lyso-derivatives. However, the stable conformation and activity of stored SMB surfactant suggest that the active helix-hairpin resists these chemical changes which otherwise may lead to surfactant inhibition.
Keywords: surfactant protein B (SP-B), Super Mini-B (SMB), captive bubble surfactometry, FTIR spectroscopy, homology modeling, surfactant lipids, lyso-phospholipids
Graphical abstract
1. Introduction
Lung surfactant is a mixture of lipids and proteins that is pivotal for normal breathing because of its ability to prevent alveolar collapse during expiration by reducing alveolar surface tension to extremely low values. Surfactant is synthesized and secreted into the alveolar fluid by alveolar type II cells and consists of approximately 80% phospholipids, 10% neutral lipids, and 10% proteins [1]. Although dipalmitoyl phosphatidylcholine (DPPC) and phosphatidylglycerol (PG) are the principal phospholipid components in lung surfactant, its biophysical activity largely depends on the presence of the hydrophobic surfactant protein B (SP-B) and, to a lesser degree, the extremely hydrophobic surfactant protein C (SP-C) [1,2]. Human SP-B is a 79 amino acid, lipid-associating monomer (MW ~8.7 kDa) that is found in the lung as a covalently linked homodimer. Each SP-B monomer consists of 4–5 α-helices with three intramolecular disulfide bridges (i.e., Cys-8 to Cys-77, Cys-11 to Cys-71 and Cys-35 to Cys-46) [3], and belongs to the saposin protein superfamily [4]. The helical bundle for SP-B is folded into two leaves, with one leaf having α-helices 1 (N-terminal helix), 5 (C-terminal helix) and 4 and the second composed of α-helices 2 and 3 [5,6].
Super Mini-B (SMB) is a 41-residue construct, based on the primary sequence, secondary structure and tertiary folding of the 79-residue SP-B, which seeks to mimic the high surfactant activity of the parent protein [6]. SMB reproduces the topology of the N- and C-terminal domains of SP-B, as it contains the N-terminal α-helix (~residues 8–25) and C-terminal α-helix (~residues 63–78) joined with a custom turn –PKGG– to form an α-helix hairpin. The oxidized SMB was shown to have two vicinal disulfide bonds (i.e., Cys-8 to Cys-77 and Cys-11 to Cys-71) that further covalently link the N- and C-terminal α-helices. Lastly, the hydrophobic N-terminal insertion sequence (i.e., residues 1–7; FPIPLPY) may insert into the lipid bilayer as an anchor for oxidized SMB attachment. Various experimental techniques were used to confirm the above structural model for oxidized SMB, including conventional 12C-FTIR spectroscopy, mass spectroscopy and Molecular Dynamics (MD) simulations in lipid mimics and lipid bilayers [6,7]. Importantly, oxidized SMB has shown excellent surface activity as single surfactant peptide in a lipid mixture that mimics the composition of native lung surfactant [6]. This synthetic lipid mixture consists of DPPC, palmitoyl-oleoyl-phosphatidylcholine (POPC) and palmitoyl-oleoyl-phosphatidylglycerol (POPG) at a weight ratio of 5:3:2 [6,8].
Intratracheal instillation of animal-derived lung surfactant extracts, which contain only polar lipids and native SP-B and SP-C, has greatly improved the survival of premature infants with neonatal respiratory distress syndrome (RDS) due to surfactant-deficiency and lung immaturity [9]. Package inserts of the porcine surfactant Curosurf® and the bovine surfactants Infasurf® and Survanta® mention a shelf life of 18 months if the vials are stored upright at a temperature of 2° to 8°C (36° to 46°F), since freezing would inactivate the dispersion and, protected from light, and state that warming and returning an unopened vial to the refrigerator is acceptable practice. However, aging and stability studies of these clinical surfactant preparations have not been reported. The latter is also true for synthetic lung surfactant formulations containing peptide mimics. For example, two commercial preparations (i.e., synthetic surfactant preparations with the α-helical SP-B mimic KL4 (Surfaxin®) and the α-helical SP-C mimic recombinant SP-Cff (Venticute®) have fallen out almost entirely because they were unstable on storage.
In this study, we measured the stability and shelf life of synthetic lung surfactant preparations consisting of 3% oxidized SMB in DPPC:POPC:POPG 5:3:2 (wt:wt:wt), in which fresh samples are compared to those stored at ~2°–8°C for 1 to 5.5 years. This surfactant preparation has been found to be highly surface active in both biophysical and animal studies [6, 10–12]. Unlike stored KL4 and recombinant SPCff formulations, captive bubble surfactometry confirmed low minimum surface tensions for fresh and stored SMB preparations. Furthermore, FTIR spectra of fresh and stored SMB formulations indicated secondary conformations compatible with SMB folding as a surface-active, α-helix-hairpin. Another contribution to SMB stability on prolonged storage may be the ‘stapling’ of the helix-hairpin with two disulfide bridges, as confirmed by both mass spectroscopy and I-TASSER homology modeling [13]. Lastly, mass spectroscopy indicated some oxidation of SMB-methionine and partial deacylation of lipids on storage. Nevertheless, the stable conformation and activity for stored SMB suggest that the active helix-hairpin resists these chemical changes by folding as a tight, globular structure in lipid.
2. Materials and Methods
2.1 Materials
HPLC grade chloroform, methanol, and acetonitrile were obtained from Fisher Scientific (Pittsburgh, PA 15275), TFA from Sigma Chemical Co (Saint Louis, MO 63103) and Sephadex LH-20 chromatography gel from Pharmacia (Uppsala, Sweden). Phospholipids were supplied by Avanti Polar Lipids (Alabaster, AL 35007) and oleic acid was obtained from NU-CHECK PREP, Inc. (Elysian, MN 56028). Sodium Dodecyl Sulfate detergent was purchased from Sigma Chemical Co (Saint Louis, MO 63103).The Super Mini-B (SMB) peptide (amino acid sequence: NH2-FPIPLPYCWLCRALIKRIQAMIPKGGRMLPQLVCRLVLRCS-COOH) was synthesized employing a standard Fmoc protocol with a Symphony Multiple Peptide Synthesizer (Protein Technologies, Inc., Tucson, AZ 85714) or a CEM Liberty microwave synthesizer (CEM Corporation, Mathews, NC 28104), cleaved-deprotected and purified using reverse phase HPLC as described previously [6]. This protocol included folding of the peptide in a structure promoting trifluorethanol-buffer solvent system to enhance the oxygen mediated disulfide linkages between Cys-8 and Cys-40 and a second linkage between Cys-11 and Cys-34 [6]. This covalently stabilized connectivity gave the peptide a helix-hairpin conformation, similar to that observed for the N-terminal and C-terminal helical domains of the saposin family of proteins [4].
2.2 Formulation and Isolation of Proteins and Lipids from Surfactant Dispersions
Peptide and lipids were formulated as lipid-peptide dispersions to have a total of 3 % by mole fraction of SMB and 35 mg of total lipid per mL of dispersion. The peptide was dissolved in 10 mL of trifluoroethanol and co-solvated with the lipid in chloroform, followed by removal of the solvents with a stream of nitrogen gas and freeze drying of the resulting lipid-peptide film to remove residual solvent. The film was then dispersed with Phosphate Buffered Saline and the sample flask containing the hydrated film was rotated for one hour at 60°C to produce a solution of multi-lamellar vesicles (MLVs) [6]. This dispersion was then stored at 4°C for various periods of time prior to structure and compositional measurements and functional surface activity determinations. In order to determine the molecular mass of peptides formulated with lipids as a function of time, the SMB was separated from the lipid using normal phase chromatography with Sephadex LH-20 [14].
2.3 Analysis of Surface Activity by Captive Bubble Surfactometry
Surface activity of the surfactant preparations was checked with a captive bubble surfactometer (CBS), which has been described in detail elsewhere [6,15]. Adsorption and surface tension lowering ability of surfactant preparations were measured at physiological cycling rate, area compression, temperature, and humidity. In brief, after inserting the bubble into the surfactant sample, adsorption of the surfactant to the bubble's air-liquid interface is monitored. After adsorption, the bubble chamber is sealed and quasi-static compression and expansion of the bubble is performed in discrete steps (at a rate of ~5% of bubble volume every 10 s) for 10 cycles. Images of the changes in bubble area are recorded during each experiment and the bubble shapes are analyzed with custom-designed software. The surface tension of the bubble is calculated on the basis of shape of the air bubble and minimum and maximum surface tension values are plotted for the first 10 cycles. We routinely insert surfactant samples of 1 µL (35 mg phospholipids/mL) into the bubble chamber with a volume of ~1.5 mL, i.e. at a concentration of ~20 µg/mL, and perform all measurements in quadruplicate.
2.4 MALDI TOF MS and HPLC Analysis of SMB
Purified SMB samples were analyzed using an AB SCIEX TOF/TOF 5800 System (Sciex, Framingham, MA 01701). Samples (~50 pmole/µL) were co-solvated with either ∝-cyano-4-hydroxycinnamic acid or sinapic acid (10 mg matrix/mL water:acetonitrile 1:1, v:v with 0.3% TFA) by mixing 24 µL of matrix solution with 1 µL of peptide solution. Two µL of this mixture was then deposited onto a metal Maldi sample plate and allowed to air dry before mass spectral measurement. Mass spectra were collected using the instrument in positive linear mode. The resulting mass spectra were analyzed with AB SCIEX Analyst and Data Explorer software.
SMB peptide samples were analyzed using a Jasco binary preparative HPLC (Jasco, Easton, MD 21601) with PU-2087 pumps and a MX-2080-32 mixer. The instrument was fitted with a Vydac 219TP DiPhenyl 250 mm by 4.6 mm reverse phase analytical column (Grace Inc., Columbia, MD 21044) and run in the analytical mode with a flow rate of 3 mL/min. The samples were chromatographed with a linear elution gradient from water to 100% acetonitrile using 0.1% trifluorethanol (v:v) as an ion pairing agent over an 1 h period. The elution of the peptide was monitored at 280 nm with a Jasco UV-2075 detector. The HPLC control and data collection was facilitated by a PC computer interfaced to the system using the Jasco ChromNAV Chromatography Data system software.
2.5 Lipid Compositional Analysis
The relative amounts of DPPC, POPC and POPG and the production of their respective lyso-phospholipid degradation derivatives in the SMB surfactant dispersions were analyzed as a function of time using LC/MS/MS (Avanti Polar Lipids, Inc., Analytical Services Division, Alabaster, AL 35007). A sample containing a total of 10 mg of combined phospholipid was dissolved in 5 ml of chloroform:methanol (v:v, 1:1) and the solution was then diluted 1:200 into the internal standard solution described below in triplicate. The relative amounts of various phospholipid molecular species in the mixture were then quantitated by an internal standard mixture containing C17:1 LPC/LPG and 17:0-14:1 PC/PG for response calibration. The internal standards and sample components were resolved by liquid chromatography of the sample prior to MS/MS detection in the negative ionization mode for the mass and fragment ions of DPPC, POPC, POPG and lyso degradants.
2.6 Attenuated-total-reflectance Fourier-Transform infrared (ATR-FTIR) of the SMB peptide in surfactant lipid dispersions
ATR-FTIR spectra were recorded at 37°C using a Bruker Vector 22 FTIR spectrometer (Pike Technologies, Fitchburg, WI 53719) with a deuterium triglyceride sulfate (DTGS) detector. The spectra were averaged over 256 scans at a gain of 4 and a resolution of 2 cm−1. The aqueous lipid-peptide solution was then transferred onto a germanium ATR crystal, and the aqueous solvent was removed by flowing nitrogen gas over the sample to produce a thick lipid-peptide film [16]. The multilayer film was then hydrated to ≥35% with deuterated water vapor in nitrogen for 1 h prior to acquiring the spectra [17]. The spectra for the SMB peptide in the lipid film were obtained by subtracting the spectrum of a peptide-free control sample from that of the peptide-bound sample. The relative amounts of α-helix, β-turn, β-sheet, or random (disordered) structures in lipid-peptide films were estimated using Fourier deconvolution (GRAMS/AI 8, version 8.0, Thermo Fisher Scientific, Waltham, MA 02451) and area of component peaks calculated using curve-fitting software (Igor Pro, version 1.6, Wavemetrics, Lake Oswego, OR 97035). FTIR frequency limits were: α-helix (1662-1645 cm−1), β-sheet (1637-1613 cm−1 and 1710-1682 cm−1), turn/bend (1682-1662 cm−1), and disordered or random (1650-1637 cm−1) [18,19].
2.7 Homology Modeling of the Structure of Oxidized SMB
The three-dimensional (3D) structure of oxidized SMB was obtained by first predicting the reduced SMB structure with a submission of the primary sequence (with four cysteines at residues 8, 11, 34, and 40) to I-TASSER V4.3 using the automated I-TASSER web service (http://zhanglab.ccmb.med.umich.edu/I-TASSER). I-TASSER is a homology algorithm that models discrete domains of the protein using multiple PDB (Protein Data Bank) depositions. The output for a predicted 3D-protein structure was a PDB file, and the accuracy of these structural predictions was estimated using parameters such as C-score, TM-score and RMSD [13,20,21]. The oxidized SMB model was next optimized by using Hyperchem 8.0 (Hypercube, Inc.; Gainesville, FL) to convert the four cysteine residues in our reduced SMB model to the known disulfide bonds at Cys-8 – Cys-40 and Cys-11 – Cys-34 [6]. Molecular graphics were rendered using Pymol Version 1.7.4.1 (Schrodinger, LLC; San Diego, CA) or MolBrowserPro 3.8-3 (Molsoft, LLC; San Diego, CA).
3. Results
3.1 In vitro surface activity
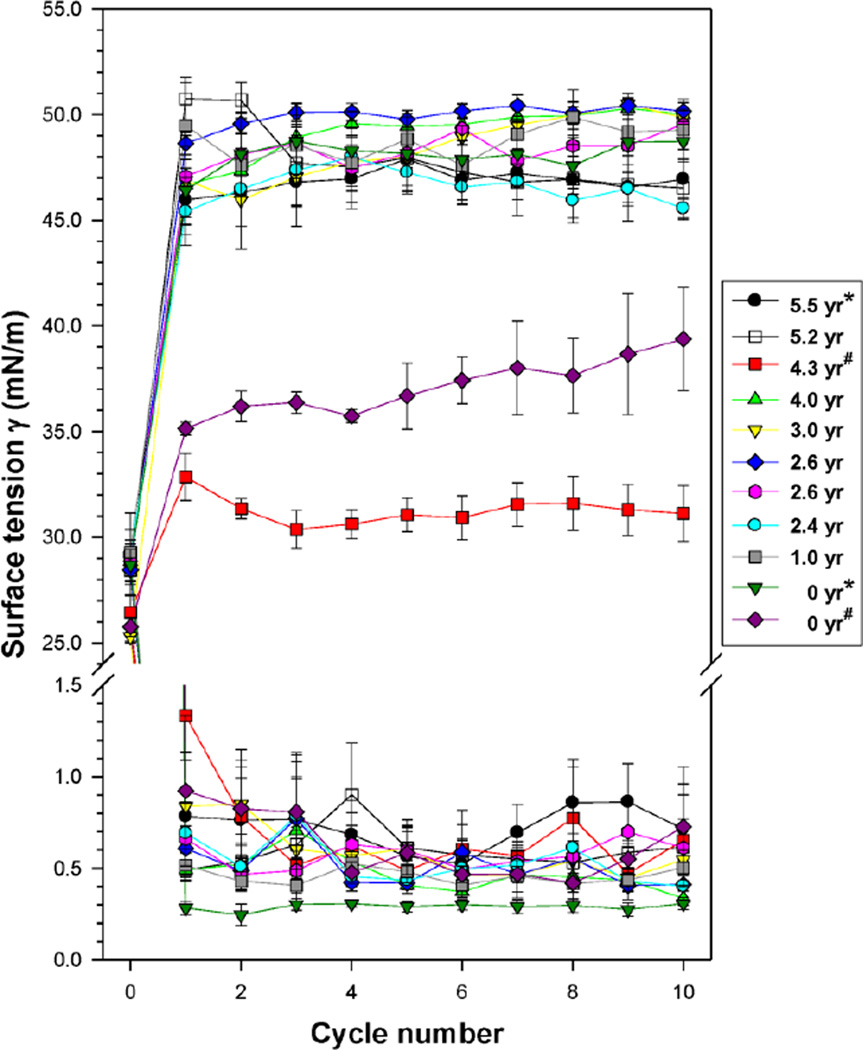
Surface activity of two ‘fresh’ SMB surfactants (0-year) and nine SMB surfactant productions that were stored at ~2–8°C for periods ranging between 1 and 5.5 years (1 of 1-year, 3 of 2-year, 1 of 3-year, 2 of 4-year and 2 of 5 year-old samples), was tested on the captive bubble surfactometer (Figure 1). The data of the two 0-year samples were obtained immediately after production of the 5.5-year and 4.3-year SMB surfactants (indicated by * and # in Figure 1). Equilibrium surface tension values right after de novo adsorption were in the 25–29 mN/m range. All surfactants tested were highly surface active: all had minimum surface tension values <1.5 mN/m from the first compression-expansion cycle onwards (Figure 1). The differences in equilibrium and minimum surface tension values were not statistically significant when analyzed by one-way analysis of variance (ANOVA). Maximum surface tension values were generally in the 45–50 mN/m range and the differences between the samples were not statistically significant, except for the 0-year surfactant sample that was re-measured at 4.3-year (p<0.02 vs all others). This SMB surfactant had maximum surface tension values <40 mN/m on both occasions with significantly lower maximum surface tension values at 4.3-year than at 0-year (p<0.05).
Figure 1.
Mean (± standard errors) of surface tension values measured by captive bubble surfactometry of 11 surfactant samples consisting of 3% Super Mini-B (SMB) in DPPC:POPC:POPG 5:3:2 wt:wt:wt aged from 0 (‘fresh’) to 5.5 years. Cycle 0 depicts surface tension values after de novo adsorption (equilibrium surface tension). Minimum surface tension values during cycles 1–10 were <1.5 mN/m in all surfactants tested and are plotted in the area below the break in the y-axis. Maximum surface tension values during cycles 1–10 are plotted in the area above the break in the y-axis. Samples marked with a * or # were measured immediately after production and again after aging. See text for details on statistical analysis. Average values ± standard errors (SEM) of 4–5 measurements per surfactant sample.
3.2 Peptide Molecular Mass as a function of time
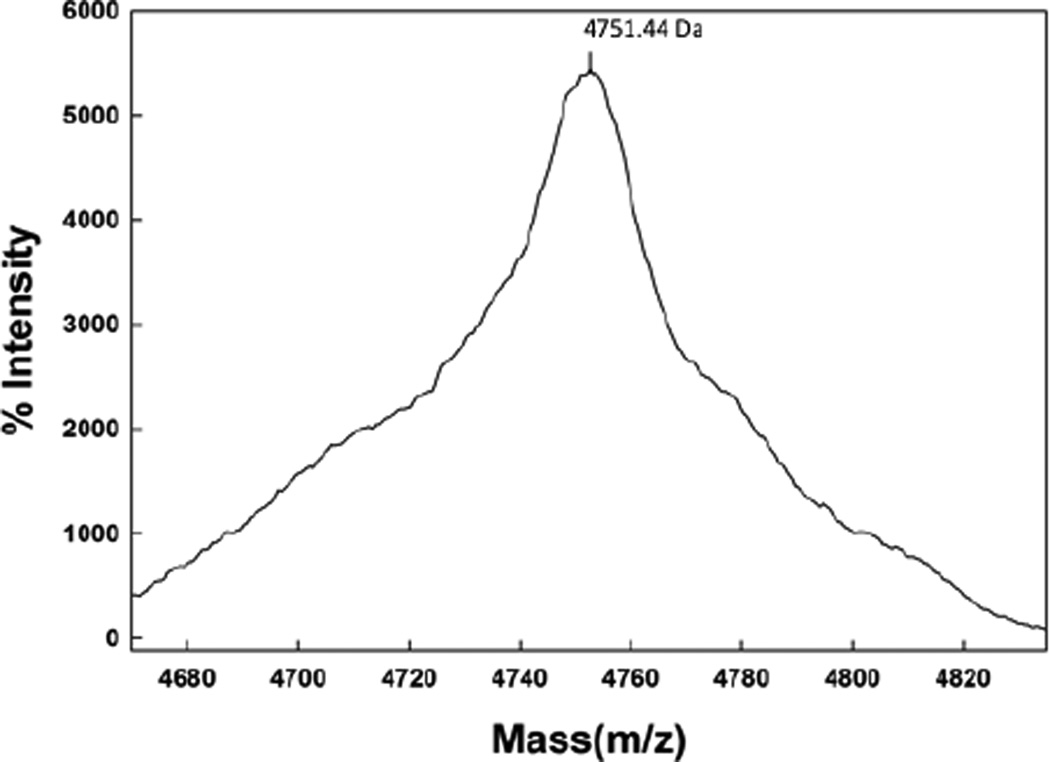
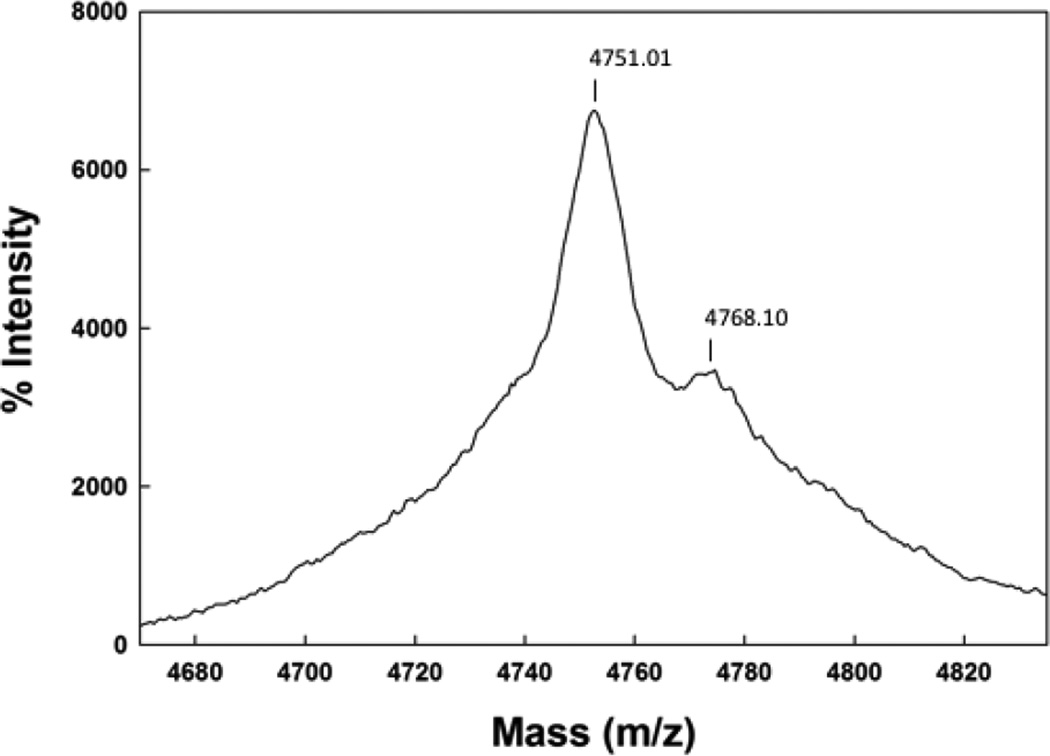
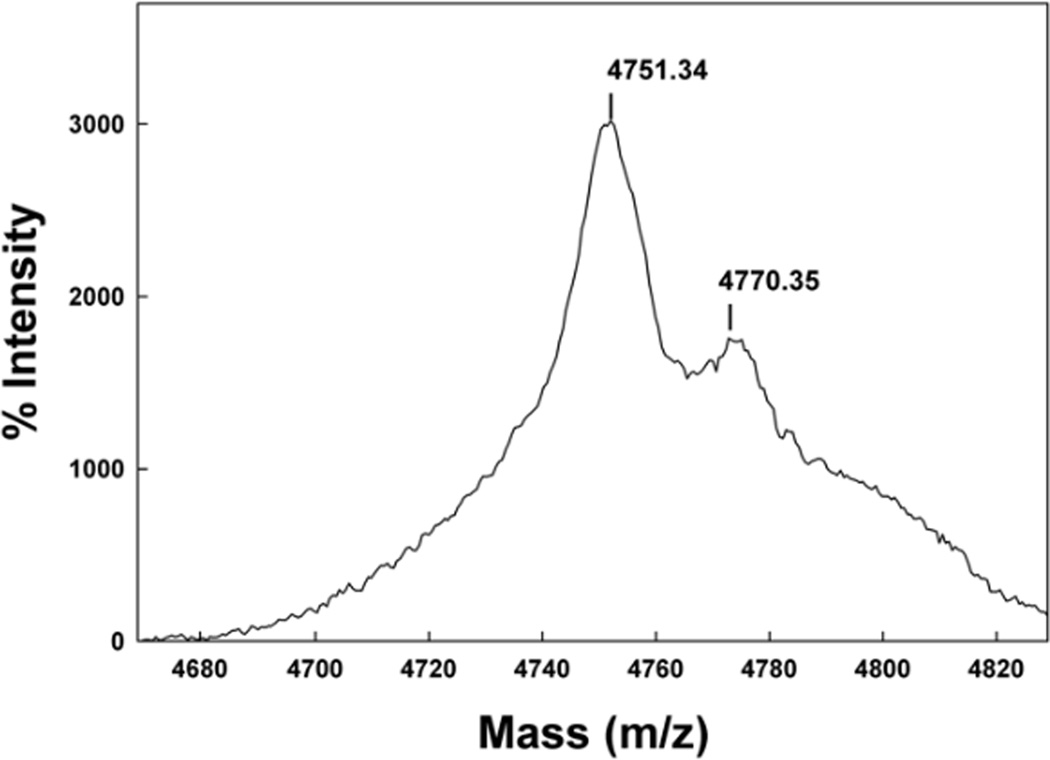
The Maldi TOF spectrum of SMB from 2 day old formulated synthetic surfactant dispersions is shown in Figure 2. The mass of 4751.44 Da from Maldi TOF analysis of these preparations is consistent with that of a disulfide linked monomeric SMB (calculated mass: 4750.97 Da). Examination of preparations stored for 1 year or more also had a dominant mass centered at 4751.01 as well as a minor mass component centered at 4768.1 Da (Figure 3). This additional minor peak suggested the addition of an oxygen atom to the peptide upon storage and most likely represents the oxidation of one of the two methionines in the peptide sequence to form the methoxide derivative. A similar pattern of methionine oxidation for SMB was observed in surfactant samples stored for a five year period (Figure 4).
Figure 2.
Maldi TOF spectra of Super Mini-B (SMB) peptide from a two day old synthetic surfactant preparation.
Figure 3.
Maldi TOF spectra of Super Mini-B (SMB) peptide from a one year old synthetic surfactant preparation.
Figure 4.
Maldi TOF spectra of Super Mini-B (SMB) peptide from a five year old synthetic surfactant preparation.
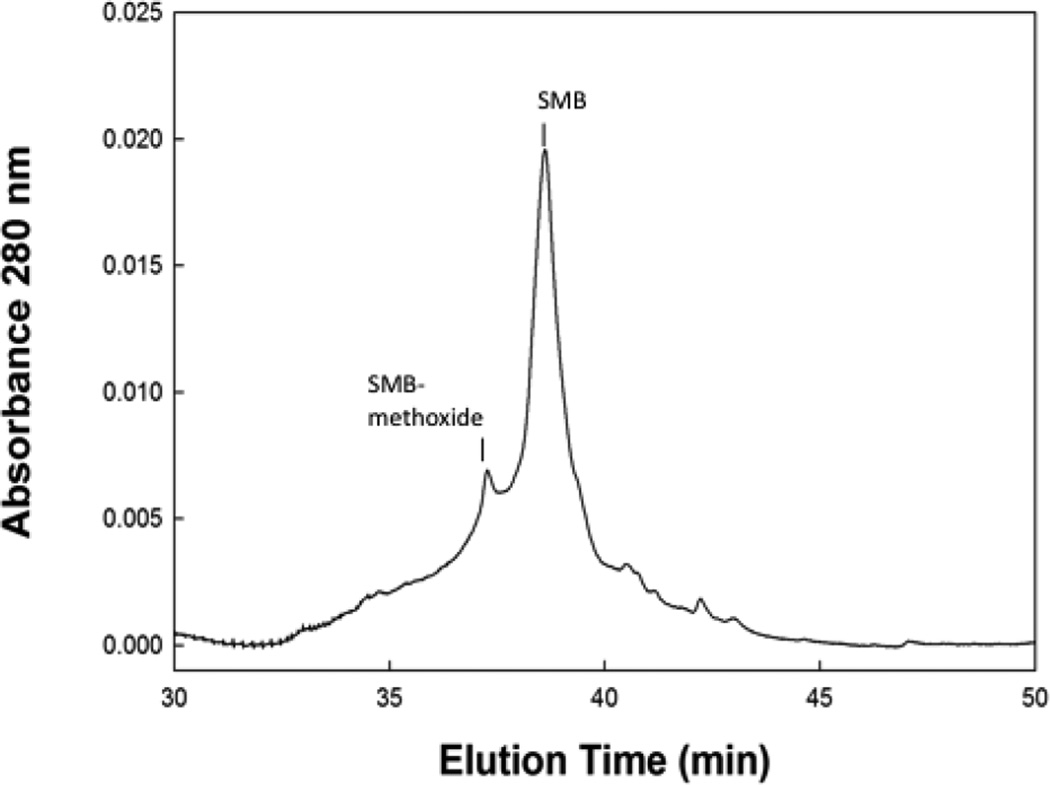
Reverse phase HPLC of stored preparation SMB peptide showed a minor peak slightly before the major parent peptide that had a mass confirming it to be the methoxide adduct (Figure 5). The elution of this component slightly before the main parent peptide is also consistent with that of the methoxide derivative having a slightly more polar character than that of the non-methionine oxidized component. Integration of the area of the 280 nm absorbance peak of the HPLC chromatogram assigned to the methoxide derivative indicated that it accounted for about 17.5% of the total peptide isolated from surfactant formulations that were stored for over one year.
Figure 5.
HPLC chromatogram of Super Mini-B (SMB) peptide from a one year old synthetic surfactant preparation.
3.3 Lipid composition of surfactant formulations as a function of time
LCMS analysis of synthetic surfactant formulations also indicated changes in the molecular species of phospholipids (Table 1) after at least one year of storage. In particular there was the formation of relatively high levels of lyso-PC in the lipid-peptide liposomal dispersion. There was also the formation of smaller amounts of lyso-PG suggesting that both phosphatidylcholine and phosphatidylglycerol underwent deacylation in the formulation with time.
Table 1.
Lipid composition as a function of time.
| Compound | 0 year, rel% | 1 year, rel% | 3 year, rel% |
|---|---|---|---|
| LysoPC | 0.3 | 8.5 | |
| POPC | 30.0 | 26.4 | 25.8 |
| DPPC | 50.0 | 60.0 | 49.3 |
| LysoPG | 0.9 | 2.5 | |
| POPG | 20.0 | 12.4 | 13.8 |
| 100.0 | 100.0 | 100.0 |
3.4 Secondary structure determination of SMB in lipid dispersions
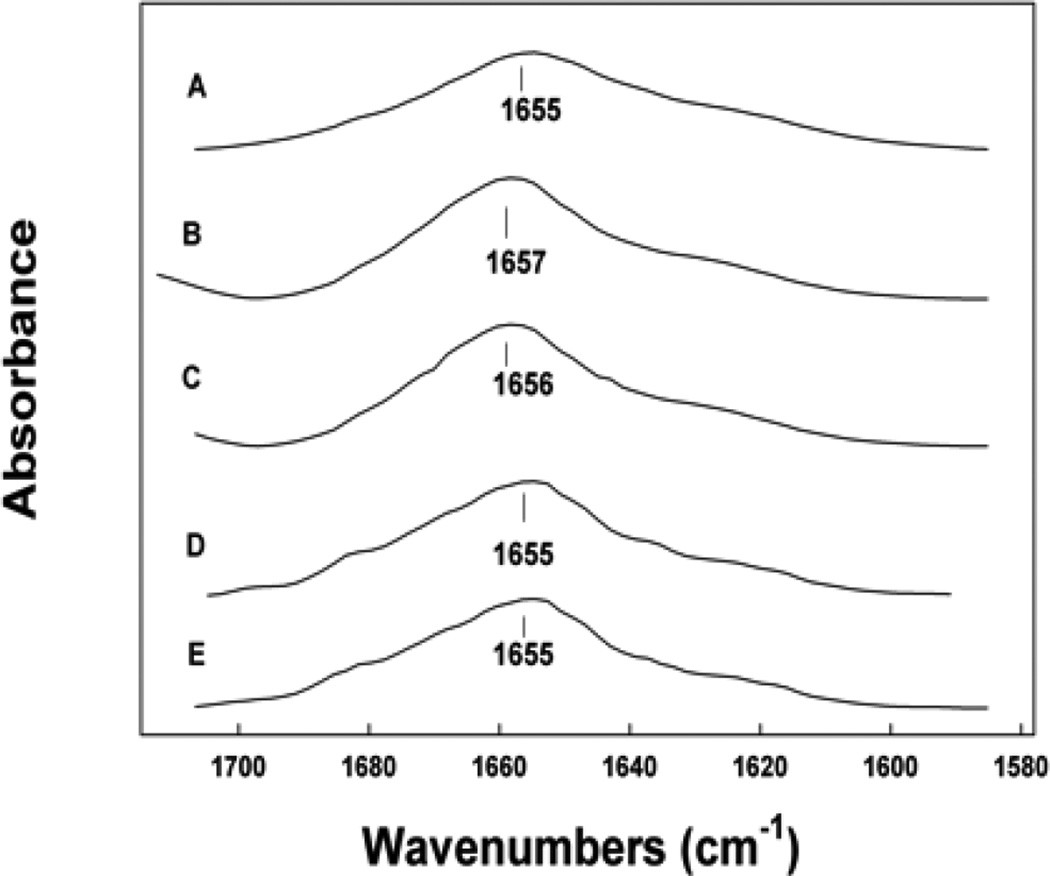
Examination of the SMB peptide in liposomal preparations over a five year period using Attenuated Total Reflectance FTIR of hydrated liposome-peptide films indicated that the peptide did not undergo significant changes in secondary conformation (Figure 6, Table 2). The peptide had a dominant alpha helical conformation in liposomal dispersions similar to that observed for the peptide in structure promoting solvents, detergent micelles and phospholipid films of lipid-peptide preparations [6]. The turn components and the beta sheet contributions associated with conformation of the peptide in surfactant lipid liposomes were also consistent over the five-year storage interval. The amounts of disordered structures were relatively small compared with other more structured conformations and also remained at the same level for the extended storage period.
Figure 6.
ATR FTIR spectra of the amide I band of Super Mini-B (SMB) peptide in synthetic surfactant lipids for various periods of storage. (A) 2 day old preparation, (B) 1 year old preparation, (C) 3 year old preparation, (D) 5 year old preparation.
Table 2.
Proportions of different components of secondary structure for SMB peptides in surfactant lipids based on FTIR spectroscopic analysis.
| Sample* | % Conformation | |||
|---|---|---|---|---|
| α-helix | turns | β-sheet | disordered | |
| SMB (2 days) | 38.5 | 24.3 | 17.7 | 19.5 |
| SMB (1 years) | 39.6 | 25.1 | 17.1 | 18.2 |
| SMB (3 years) | 38.8 | 25.6 | 16.9 | 18.7 |
| SMB (5 years) | 38.7 | 24.1 | 19.2 | 18.0 |
Peptides in MLV dried films hydrated with D2O were analyzed for secondary conformation based on secondary structural analysis using spectral deconvolution of the ATR-FTIR spectra of the peptide amide I band (Methods). Percent conformation represents average value of 8 measurements with SEM of ± 0.33 or better.
To further understand the possible correlation between SMB helical propensity and the membrane like environment the peptide was interacting with, we used anionic micelles and lipid ensembles with highly elevated levels of lyso and fatty acid derivatives. In SDS micelles the helix hairpin peptide still assumed the dominant alpha helical conformation (Figure 6, Table 3) similar to that observed in synthetic surfactant lipid dispersions. Similar high levels of SMB alpha helical conformations were observed for peptide in lyso phospholipid micelles (Figure 7, Table 3), indicating that even in micelles with anionic charge and high positive curvature the helix hairpin assumed the same secondary conformation.
Table 3.
Proportions of different components of secondary structure for SMB peptides in micelles and surfactant lipids based on FTIR spectroscopic analysis.
| Sample* | % Conformation | |||
|---|---|---|---|---|
| α-helix | turns | β-sheet | disordered | |
| SMB-SDS | 44.5 | 25.2 | 5.7 | 24.6 |
| SMB-LysoPG | 46.9 | 22.9 | 13.1 | 17.1 |
| SMB-DPPC- LysoPG | 46.1 | 25.1 | 14.2 | 14.7 |
| SMB-DPPC- oleic acid | 44.7 | 20.5 | 14.6 | 20.2 |
| SMB-DPPC- LysoPG- oleic acid | 40.7 | 30.0 | 10.8 | 18.5 |
Peptides in MLV dried films hydrated with D2O were analyzed for secondary conformation based on secondary structural analysis using spectral deconvolution of the ATR-FTIR spectra of the peptide amide I band (Methods). Percent conformation represents average value of 5 measurements with SEM of ± 0.38 or better.
Figure 7.
ATR FTIR spectra of the amide I band of Super Mini-B (SMB) peptide in micelles and synthetic surfactant lipids. (A) SMB in SDS micelles, peptide to lipid ratio 1:50, mole:mole, (B) SMB in Miristoyl Lyso-PG micelles, peptide to lipid 1:60, mole:mole, (C) SMB in DPPC: Miristoyl Lyso-PG MLVs, 1:5:5, mole:mole, (D) SMB in DPPC: oleic acid, 1:5:5, mole:mole MLVs, (E) SMB in DPPC: Miristoyl Lyso-PG:oleic acid, 1:5:2.5:2.5, mole:mole MLVs.
Examination of multilayer vesicles (MLVs) containing elevated levels of the deacylation products from long term storage of synthetic surfactant dispersions also confirmed the stability of the peptide in multiple environments (Figure 7, Table 3). In MLVs of DPPC – lyso-PG, DPPC – oleic acid or DPPC with high concentrations of lyso-PG and oleic acid in equal mole ratios, the peptide maintained the alpha helical – hair pin conformations associated with other preparations.
3.5 Homology Modeling the Structure of the Oxidized SMB Peptide
Although the above 12C-FTIR spectral experiments are useful for assessing secondary structures averaged over the whole peptide, they cannot indicate the conformations of individual amino-acids. One method to address this deficiency has been to find the 3D-structure of a homologous protein and then build a comparable 3D-model based on this template structure and the primary sequence of the target protein [22]. In more recent work [13,21], I-TASSER was developed as a homology algorithm that models distinct regions of the protein using multiple PDB (Protein Data Bank) depositions. I-TASSER is an automated pipeline for structure predictions using multiple threading alignments and simulations of iterative assemblies and has successfully forecast a range of protein structures [13,21,23,24].
In the present study, the three-dimensional (3D) structure of the oxidized SMB peptide was predicted by our first submitting the reduced SMB sequence (with four cysteine residues with thiol groups) to the I-TASSER web service (http://zhanglab.ccmb.med.umich.edu/I-TASSER). Three distinct models for reduced SMB were obtained, and Model 1 with the highest C-score was selected (see Fig. 8). The accuracy for Model 1 was estimated from the following parameters: C-score of 0.04, TM-score of 0.72 ± 0.11 and RMSD of 2.2 ± 1.7 Å. C-score is a confidence score for evaluating the quality of I-TASSER models (between −5 to 2), with elevated values indicating a model with high confidence [13,21]. TM-score is a scale for quantifying the similarity between two structures, with scores greater than 0.50 signifying a model of correct topology and scores less than 0.17 implying random similarity [13,20,21,25]. Moreover, RMSD (i.e. root mean square deviation) is an average distance of all residue pairs in the two structures. The high C- and TM-scores, together with the low RMSD, indicate that Model 1 provides accurate estimates of the secondary and tertiary structures for reduced SMB. The oxidized SMB model was next made from Model 1 by forming the disulfide bridges between Cys-8 – Cys-40 and Cys-11 – Cys-34. Here, Hyperchem 8.0 minimized the respective lengths between the Cys – Cys residues so that the paired -S – S- distances (i.e., 1.84 Å for Cys-8 – Cys-40 and 2.04 Å for Cys-11 – Cys-34) are similar to the experimental values reported in other saposins [5]. Because Hyperchem 8.0 optimized only the Cys residues for the formation of disulfide bridges, both the main chains and other sidechains were largely unaffected.
Figure 8.
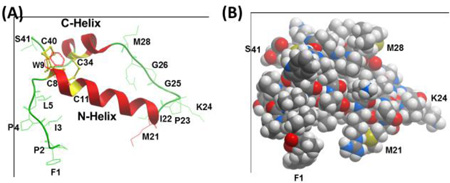
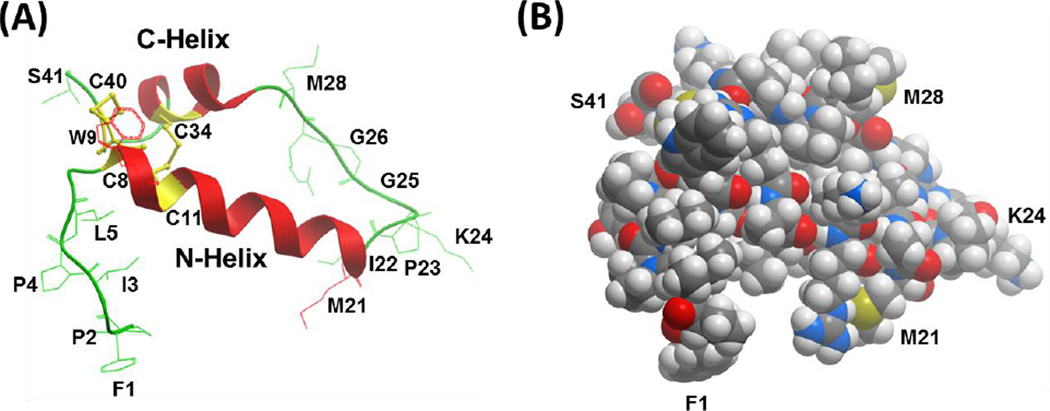
Molecular graphics representations of the homology model for oxidized Super Mini-B (SMB), in ribbon (A) or solid CPK (B) formats. The 3D-structure of oxidized SMB was predicted by first using I-TASSER V4.3 to calculate the best model (i.e., Model 1) for reduced SMB with four cysteines at C8, C11, C34, and C40. The oxidized SMB model was next determined by using Hyperchem 8.0 to convert the four cysteine residues in reduced SMB to disulfide bonds at C8 – C40 and C11 – C34. (A) The ribbon model for oxidized SMB predicts a C-terminal α-helix (i.e., red C-Helix, residues P30-L36) connected to an N-terminal α-helix (i.e., red N-Helix; residues C8-M21) via a coil (green residues I22-L29), which adopts a helix hairpin conformation. The homology model also forecasts a flexible coil for the N-terminal insertion sequence (F1 – Y7), which allows this segment to interact partially with the N-helix. One of the two methionines (i.e., M21, M28) may oxidize to methoxide under our storage conditions. (B) The CPK homology model for oxidized SMB shows the space-filling representation in the same orientation as the ribbon structure to the left (see labeled F1, M21, K24, M28 and S41 residues). The color key for the atoms is C (gray), O (red), N (blue), S (yellow) and H (white). Graphical representations of PDB files were rendered with Molsoft MolBrowserPro 3.8-3.
The above homology model for oxidized SMB (see ribbon representation in Fig. 8A) predicts a C-terminal α-helix (residues P30-L36) connected to an N-terminal α-helix (residues C8-M21) via a coil (residues I22-L29), which adopts a helix hairpin conformation [26,27]. Here, the putative helix hairpin, or helix-hairpin-helix, executes a reverse turn after 8-residues with the I22-G25 component being the most prominent. Similar to previous helix hairpins [27], neural network and homology analysis of the SMB sequence [28,29] indicated high β-turn propensities for M21 to G25, permitting close interactions between the hydrophobic interfaces of the nearly antiparallel N- and C- helices. The homology model also forecasts a flexible coil for the N-terminal insertion sequence (F1 – Y7), which allows this hydrophobic segment to interact partially with the N-helix. Additionally, one of the two methionines (i.e., M21, M28) in Fig. 8A may be oxidized to methoxide under our storage conditions. Lastly, the space-filling I-TASSER model of oxidized SMB in Fig. 8B shows that the folding of SMB into a disulfide cross-linked, helix hairpin produces a compact globular protein, exhibiting a core of hydrophobic residues and a surface of aqueous-exposed and polar residues.
4. Discussion
In this study, we tested the stability and function of SMB surfactant preparations that had been stored in our cold room for periods ranging from 1 to up to 5 years. Captive bubble surfactometry demonstrated persistent high in vitro surface activity of these samples in comparison with a fresh sample. FTIR spectroscopy of the SMB peptide showed that extended storage did not affect the secondary structure of SMB in surfactant liposomes, although mass spectroscopy indicated some oxidation of methionine residues with time. However, some oxidation of the sulfur of methionine is also observed in SP-B from fresh natural lung surfactant [30] and we found that methionine oxidation did not affect peptide structure and only a minor change in polarity. The synthetic surfactant lipids (DPPC, POPC and POPG) were affected by deacylation leading to the formation of Lyso-Palmitoyl PC and Lyso-Palmitoyl PG over the five year storage period. However, the structure and activity of SMB was not altered by the presence of high levels phospholipid lyso-derivatives in the stored samples, suggesting that the helix – hairpin derivative resists the surface activity inhibition of these lipid components in synthetic surfactant dispersions. This is in line with previous work by our group on surfactant inhibition by lyso-derivatives [24]. The relative stability of the SMB structure in the ageing samples was confirmed by extended molecular dynamics simulations in a lipid environment and in micelles of anionic detergent and lysophosphatidyl phosphoglycerol (Waring et al., unpublished observations).
Liquid lung surfactant preparations are inherently more sensitive to oxidative damage than dry powder preparations, but exact data on ageing of clinical, i.e. animal-derived, lung surfactants with native SP-B and SP-C have not been published except that package inserts mention a shelf-life of 18 months during storage at a temperature of 2° to 8°C. Oxidation of SMB by exposure to the air pollutant ozone negatively affects its surface activity, probably by partial unfolding that impairs its interaction with negatively charged PG lipids at the air-water interface [31]. Exposure of mixed monolayers composed of POPC and DPPC to ozone does not lead to a loss of DPPC from the air-water interface, but surface activity changes significantly secondary to oxidative damage of the unsaturated POPC [32]. Similar findings have been reported for the shorter SP-B mimic SP-B1–25 in unsaturated 1-palmitoyl-2-oleoyl-sn-glycerol [33]. Both components were affected by exposure to ozone resulting in reduced surface activity at the air-water interface. On the contrary, 60 hours of hyperoxia (95% oxygen) in rats does not lead to a dysfunction of native surfactant proteins, and although PG lipids are susceptible to oxidation this leads only to a minor disturbance in surface activity [34]. We recently demonstrated that exposure of synthetic lung surfactant consisting of SMB-DATK peptide, a SMB analogue with a designer loop, and DPPC:POPC:POPG 5:3:2 was not sensitive to inhibition by Lyso-PC in concentrations of up to 20% [24], i.e. far higher than the concentration of lyso lipids here.
The two disulfide bridges in oxidized SMB (at Cys-8 – Cys-40 and Cys-11 – Cys-34) are likely to play critical roles in the long-term structural and functional stability reported here for this protein. Long-term storage of SMB surfactant preparations for a year or more showed not only elevated in vitro activities but also higher α-helix and lower secondary structural components (i.e., loop-turn, β-sheet and random) that are a hallmark of highly active SMB. Because the two disulfide bridges similarly persist on long-term storage, it is not untoward to speculate that the disulfide bonds are involved in these structural or functional properties, or both. As noted in Fig. 8A, the homology ribbon model indicates that SMB folds as a helix-hairpin, with disulfides at Cys-8 – Cys-40 and Cys-11 – Cys-34 bridging the N- and C-α-helices. The disulfide bonds will probably constrain SMB by bringing the N- and C-α-helices closer together, shifting SMB to its final helix-hairpin configuration. In effect, a disulfide bond destabilizes the unfolded conformer of the peptide by reducing its entropy [35]. The homology space-filling model for SMB in Fig. 7B demonstrates a globular protein, in which the two disulfides are embedded with other hydrophobic residues in the interior and hydrophilic groups face the exterior. Here, the disulfide bonds may act as a hydrophobic core around which other hydrophobic residues cluster. By linking the N- and C-α-helices together, the two disulfide bonds raise the local concentration of protein residues while reducing the local concentration of water. Because water molecules disrupt secondary structure by attacking amide-amide hydrogen bonds, the remarkable SMB stability to long-term storage may be principally due to water exclusion from the hydrophobic interior. In support of this hypothesis, preliminary 499-nsec MD simulations of SMB in surfactant lipid bilayers [i.e., DPPC:POPC:DOPG] showed the absence of any water within oxidized SMB (Waring et al., unpublished observations). Future experiments are planned to study the structural and functional roles for disulfides using the reduced SMB peptide that lacks cystine residues [24].
Highlights.
Super Mini-B (SMB) is a disulfide-linked helix-hairpin with high surfactant activity
Activity and conformation of SMB in lipid were not changed by storage (4°C for 5 years)
This stability is due to SMB folding as a helix-hairpin 'stapled' with two disulfides
Some oxidation of the SMB methionine and partial deacylation of lipids occurred on storage
SMB resisted chemical inhibition by adopting a tight, globular structure in lipids
Acknowledgments
This work was supported by the National Institutes of Health [grant R01HL092158] and the Bill & Melinda Gates Foundation, Seattle, WA [grant OPP1112090].
Abbreviations
- ATR-FTIR
Attenuated-total-reflectance Fourier-Transform infrared
- CBS
captive bubble surfactometer
- DPPC
dipalmitoyl-phosphatidylcholine
- DTGS
deuterium triglyceride sulfate
- MD
molecular dynamics
- MLVs
multi-lamellar vesicles
- PBS
phosphate buffered saline
- POPC
palmitoyl-oleoyl-phosphatidylcholine
- POPG
palmitoyl-oleoyl-phosphatidylglycerol
- RDS
respiratory distress syndrome
- SMB
Super Mini-B
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
Author contributions
A.W. and F.W. designed the research. A.W., F.W. and G.F. provided materials. A.W., M.G., L.G. and F.W. performed the research. A.W., L.G. and F.W. analyzed the data and wrote the manuscript. All authors discussed the results and approved the manuscript.
References
- 1.Goerke J. Pulmonary surfactant: functions and molecular composition. Biochim. Biophys. Acta. 1998;1408:79–89. doi: 10.1016/s0925-4439(98)00060-x. [DOI] [PubMed] [Google Scholar]
- 2.Walther FJ, Waring AJ, Sherman MA, Zasadzinski JA, Gordon LM. Hydrophobic surfactant proteins and their analogues. Neonatology. 2007;91:303–310. doi: 10.1159/000101346. [DOI] [PubMed] [Google Scholar]
- 3.Johansson J, Jörnvall H, Curstedt T. Human surfactant polypeptide SP-B. Disulfide bridges, C-terminal end, and peptide analysis of the airway form. FEBS Lett. 1992;301:165–167. doi: 10.1016/0014-5793(92)81239-i. [DOI] [PubMed] [Google Scholar]
- 4.Munford RS, Sheppard PO, O’Hara PJ. Saposin-like proteins (SAPLIP) carry out diverse functions on a common backbone structure. J. Lipid Res. 1995;36:1653–1663. [PubMed] [Google Scholar]
- 5.Bruhn H. A short guided tour through functional and structural features of saposin-like proteins. Biochem. J. 2005;389:249–257. doi: 10.1042/BJ20050051. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Walther FJ, Waring AJ, Hernadez-Juviel JM, Gordon LM, Wang Z, Jung C-L, Ruchala P, Clark AP, Smith WM, Sharma S, Notter RH. Critical structural and functional roles for the N-terminal insertion sequence in surfactant protein B analogs. PLoS One. 2010;5:e8672. doi: 10.1371/journal.pone.0008672. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Schwan AL, Singh SP, Davy JA, Waring AJ, Gordon LM, Walther FJ, Wang Z, Notter RH. Synthesis and activity of a novel diether phosphonoglycerol in phospholipase-resistant synthetic lipid:peptide lung surfactants. Medchemcomm. 2011;2:1167–1173. doi: 10.1039/C1MD00206F. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Walther FJ, Hernandez-Juviel J, Gordon LM, Waring AJ, Stenger P, Zasadzinski JA. Comparison of three lipid formulations for synthetic surfactant with a surfactant protein B analog. Exp. Lung Res. 2005;31:563–579. doi: 10.1080/019021490951531. [DOI] [PubMed] [Google Scholar]
- 9.Polin RA, Carlo WA. Committee on Fetus and Newborn; American Academy of Pediatrics. Surfactant replacement therapy for preterm and term neonates with respiratory distress. Pediatrics. 2014;133:156–163. doi: 10.1542/peds.2013-3443. [DOI] [PubMed] [Google Scholar]
- 10.Walther FJ, Hernández-Juviel JM, Gordon LM, Waring AJ. Synthetic surfactant containing SP-B and SP-C mimics is superior to single-peptide formulations in rabbits with chemical acute lung injury. PeerJ. 2014;2:e393. doi: 10.7717/peerj.393. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Sharifahmadian M, Sarker M, Palleboina D, Waring AJ, Walther FJ, Morrow MR, Booth V. Role of the N-terminal seven residues of surfactant protein B (SP-B) PLoS One. 2013;8:e72821. doi: 10.1371/journal.pone.0072821. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Gupta R, Hernández-Juviel JM, Waring AJ, Walther FJ. Synthetic lung surfactant reduces alveolar-capillary protein leakage in surfactant-deficient rabbits. Exp. Lung Res. 2015;41:293–299. doi: 10.3109/01902148.2015.1024354. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Zhang Y. I-TASSER server for protein 3D structure prediction. BMC Bioinformatics. 2008;9:40. doi: 10.1186/1471-2105-9-40. 2008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Baatz JE, Zou Y, Cox JT, Wang Z, Notter RH. High-yield purification of lung surfactant proteins SP-B and SP-C and the effects on surface activity. Protein Expr. Purif. 2001;23:180–190. doi: 10.1006/prep.2001.1492. [DOI] [PubMed] [Google Scholar]
- 15.Schürch S, Green FH, Bachofen H. Formation and structure of surface films: captive bubble surfactometry. Biochim Biophys Acta. 1998;1408:180–202. doi: 10.1016/s0925-4439(98)00067-2. [DOI] [PubMed] [Google Scholar]
- 16.Walther FJ, Waring AJ, Hernandez-Juviel JM, Ruchala P, Wang Z, Notter RH, Gordon LM. Surfactant protein C peptides with salt-bridges (“ion-locks”) promote high surfactant activities by mimicking the α-helix and membrane topography of the native protein. PeerJ. 2014;2:e485. doi: 10.7717/peerj.485. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Yamaguchi S, Hong T, Waring A, Lehrer RI, Hong M. Solid-state NMR investigations of peptide-lipid interaction and orientation of a beta-sheet antimicrobial peptide, protegrin. Biochemistry. 2002;41:9852–9862. doi: 10.1021/bi0257991. [DOI] [PubMed] [Google Scholar]
- 18.Kauppinen JK, Moffatt DJ, Mantsch HH, Cameron DG. Fourier self-deconvolution: a method for resolving intrinsically overlapped bands. Appl. Spectrosc. 1981;35:271–276. [Google Scholar]
- 19.Byler DM, Susi H. Examination of the secondary structure of protein by deconvolved FTIR spectra. Biopolymers. 1986;25:469–487. doi: 10.1002/bip.360250307. [DOI] [PubMed] [Google Scholar]
- 20.Zhang Y, Skolnick J. Scoring function for automated assessment of protein structure template quality. Proteins. 2004;57:702–710. doi: 10.1002/prot.20264. [DOI] [PubMed] [Google Scholar]
- 21.Yang J, Yan R, Roy A, Xu D, Poisson J, Zhang Y. The I-TASSER Suite: protein structure and function prediction. Nat. Methods. 2015;12:7–8. doi: 10.1038/nmeth.3213. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Liang J, Naveed H, Jimenez-Morales D, Adamian L, Lin M. Computational studies of membrane proteins: Models and predictions for biological understanding. Biochim. Biophys. Acta. 2012;1818:927–941. doi: 10.1016/j.bbamem.2011.09.026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Saxena R, Singh R. MALDI-TOF MS and CD spectral analysis for identification and structure prediction of a purified, novel, organic solvent stable, fibrinolytic metalloprotease from Bacillus cereus B80. Biomed. Res. Int. 2015;2015:527015. doi: 10.1155/2015/527015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Notter RH, Wang Z, Walther FJ. Activity and biophysical inhibition resistance of a novel synthetic lung surfactant containing Super-Mini-B DATK peptide. PeerJ. 2016;4:e1528. doi: 10.7717/peerj.1528. 2016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Zhang Y, Skolnick J. TM-align: a protein structure alignment algorithm based on the TM-score. Nucleic Acids Res. 2005;33:2302–2309. doi: 10.1093/nar/gki524. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Engelman DM, Steitz TA. The spontaneous insertion of proteins into and across membranes: the helical hairpin hypothesis. Cell. 1981;23:411–422. doi: 10.1016/0092-8674(81)90136-7. [DOI] [PubMed] [Google Scholar]
- 27.Thayer MM, Ahern H, Xing D, Cunningham RP, Tainer JA. Novel DNA binding motifs in the DNA repair enzyme endonuclease III crystal structure. EMBO J. 1995;14:4108–4120. doi: 10.1002/j.1460-2075.1995.tb00083.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Kaur H, Raghava GP. Prediction of beta-turns in proteins from multiple alignment using neural network. Protein Sci. 2003;12:627–634. doi: 10.1110/ps.0228903. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Kaur H, Raghava GP. A neural network method for prediction of beta-turn types in proteins using evolutionary information. Bioinformatics. 2004;20:2751–2758. doi: 10.1093/bioinformatics/bth322. [DOI] [PubMed] [Google Scholar]
- 30.Manzanares D, Rodriguez-Capote K, Liu S, Haines T, Ramos Y, Zhao L, Doherty-Kirby A, Lajoie G, Possmayer F. Modification of tryptophan and methionine residues is implicated in the oxidative inactivation of surfactant protein B. Biochemistry. 2007;46:5604–5615. doi: 10.1021/bi062304p. [DOI] [PubMed] [Google Scholar]
- 31.Hemming JM, Hughes BR, Rennie AR, Tomas S, Campbell RA, Hughes AV, Arnold T, Botchway SW, Thompson KC. Environmental pollutant ozone causes damage to lung surfactant protein B (SP-B) Biochemistry. 2015;54:5185–5197. doi: 10.1021/acs.biochem.5b00308. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Thompson KC, Jones SH, Rennie AR, King MD, Ward AD, Hughes BR, Lucas CO, Campbell RA, Hughes AV. Degradation and rearrangement of a lung surfactant lipid at the air-water interface during exposure to the pollutant gas ozone. Langmuir. 2013;29:4594–4602. doi: 10.1021/la304312y. [DOI] [PubMed] [Google Scholar]
- 33.Kim HI, Kim H, Shin YS, Beegle LW, Jang SS, Neidholdt EL, Goddard WA, Heath JR, Kanik I, Beauchamp JL. Interfacial reactions of ozone with surfactant protein B in a model lung surfactant system. J. Am. Chem. Soc. 2010;132:2254–2263. doi: 10.1021/ja908477w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Pace PW, Yao LJ, Wilson JX, Possmayer F, Veldhuizen RA, Lewis JF. The effects of hyperoxia exposure on lung function and pulmonary surfactant in a rat model of acute lung injury. Exp. Lung Res. 2009;35:380–398. doi: 10.1080/01902140902745166. [DOI] [PubMed] [Google Scholar]
- 35.Futami J, Tada H, Seno M, Ishikami S, Yamada H. Stabilization of human RNase 1 by introduction of a disulfide bond between residues 4 and 118. J. Biochem. 2000;128:245–250. doi: 10.1093/oxfordjournals.jbchem.a022747. [DOI] [PubMed] [Google Scholar]