Fig. 1.
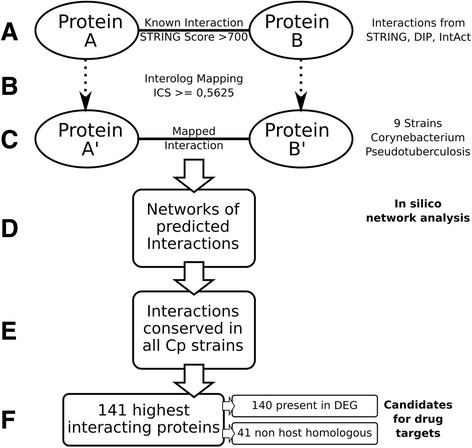
Overview of the workflow utilized in this manuscript. a We extracted known interactions from the STRING, DIP, and IntAct databases. b For each interaction we searched for conserved counterparts in the nine Cp strains and mapped the interaction in case an ICS score of 0.5625 or larger was achieved (c). The mapped interactions form the candidate networks for the Cp strains (d). For the further investigation we only used those interactions which are present in all nine strains (e) and extracted the top 15 % proteins with the highest interaction degree (f) as they are most likely to be essential proteins. Of the selected 181 proteins were 180 indeed present in the DEG database and furthermore, 41 of those have no homologous counterpart in the host organisms and thus are promising potential drug targets.
