Fig. 2.
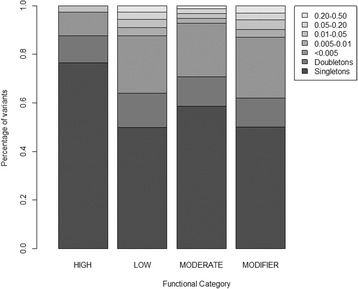
Minor allele frequency distribution of observed single-nucleotide variants (SNVs). SNVs are categorized by functional impact (high, moderate, low, or modifier) as predicted by the SnpEff algorithm [43]. High (n = 81) is defined as disruptive impact SNVs, moderate (n = 1983) as nonsynonymous coding SNVs, low (n = 1374) as synonymous coding SNVs, and modifier (n = 134,092) as noncoding SNVs
