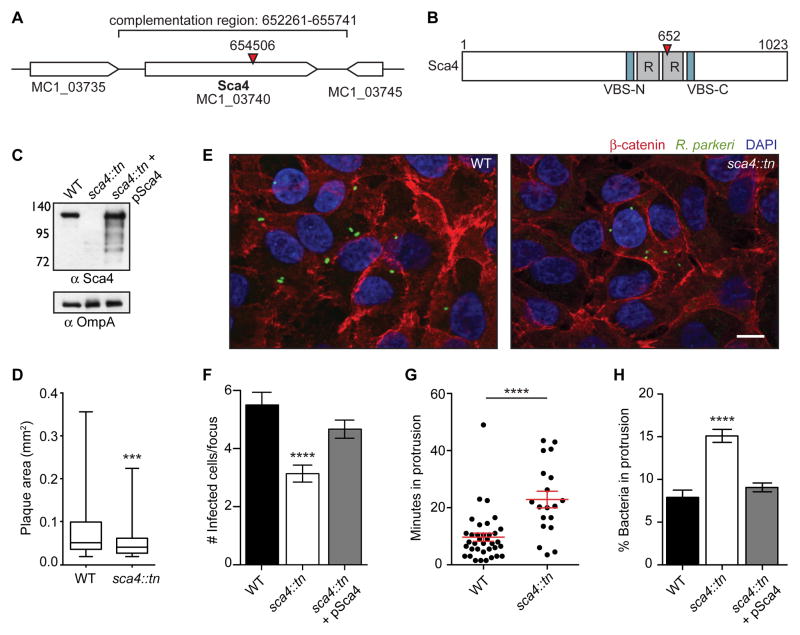
Figure 2. Transposon mutagenesis of R. parkeri sca4 leads to a defect in cell-to-cell spread.
(A) R. parkeri chromosome region containing the sca4 gene (red arrowhead, tn insertion; bracket, region used for complementation). (B) Sca4 protein schematic indicating the VBSs, predicted antigenic repeats (R) and tn insertion at residue 652 (red arrowhead). (C) Western blot of purified R. parkeri strains, WT (Rp-GFP), sca4::tn and sca4::tn + pSca4 (pRAM18dSGA-Sca4). (OmpA, loading control) (D) Plaque areas in cell monolayers infected with WT and sca4::tn R. parkeri. (E) Images of infectious foci formed by WT (Rp-GFP) and sca4::tn strains. (Red, β-catenin; green, R. parkeri; blue, DAPI). Scale bar, 10 μm. (F) Infectious focus sizes formed by WT (Rp-GFP), sca4::tn and sca4::tn + pSca4 strains. (G) Average time each bacterium spent in a protrusion before resolution into a vesicle (WT data are from Figure 1C). (H) Percentage of bacteria within a protrusion. Data for (D) and (F–H) are mean ± SEM, unpaired T-test: *** p < 0.001 **** p < 0.0001 relative to WT. See Figure S1.