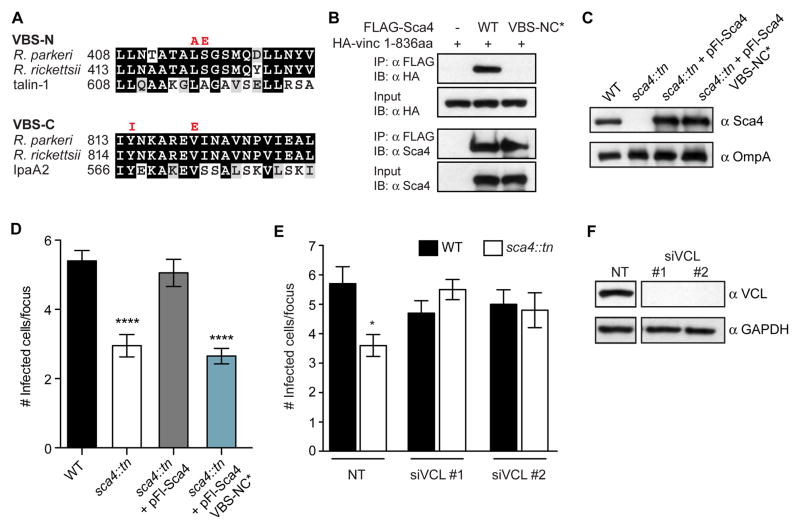
Figure 4. Sca4 targets vinculin to promote spread.
(A) Sequence alignments of the indicated VBSs (black, identical residues; grey, similar residues). VBS-NC* mutant amino acid substitutions are indicated in red. (B) Co-IP between FLAG-tagged Sca4 (WT and VBS-NC*) and the 1–836aa truncation of vinculin. IP, immunoprecipitate; IB, immunoblot. (C) Western blot showing expression levels of Sca4 in indicated purified R. parkeri strains. (D) Infectious focus size formed by WT (Rp-GFP), sca4::tn, sca4::tn + pFl-Sca4 (pRAM18dSGA-Sca4pr-Flag-Sca4) and sca4::tn + pFl-Sca4 VBS-NC* strains. Mean ± SEM, unpaired T-test: **** p < 0.0001 relative to WT. (E) Infectious focus size after reverse transfection with control siRNA (nontarget, NT) or vinculin-specific siRNAs (siVCL #1, #2) and infection with WT (Rp-GFP) or sca4::tn. Mean ± SEM, unpaired T-test: **** p < 0.0001 relative to WT + NT. (F) Western blot of A549 lysates from (E) (GAPDH, loading control). Space in blots indicates deletion of irrelevant lane.