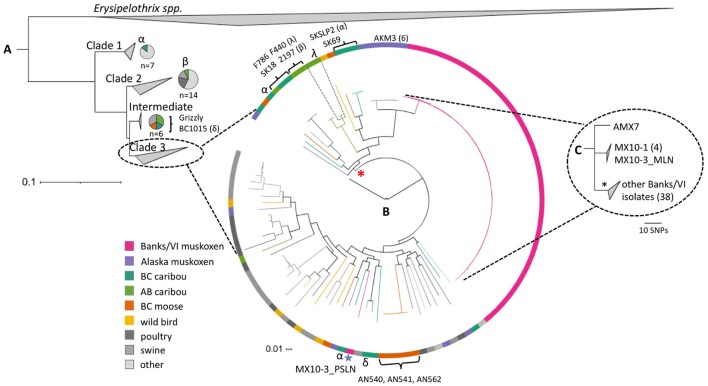
Figure 4.
The diversity of Erysipelothrix rhusiopathiae in North American wild ungulates, shown within the broader global population structure. (A) The phylogenetic tree of the three major clades (collapsed) generated in PhyloPhlAn using >400 conserved bacterial protein sequences, and rooted to other Erysipelothrix spp. (E. tonsillarum and E. sp. strain 2). It comprises one hundred-fifty E. rhusiopathiae isolates, including sequences available on GenBank and isolates previously described (Forde et al., 2016). Specific isolates named in this figure are described in Table 2. The species breakdown within Clades 1, 2 and intermediate is shown using pie-charts. (B) The relationship among isolates within the dominant Clade 3, shown with a circularized maximum likelihood (ML) tree rooted to the Fujisawa reference genome (intermediate clade). This tree is based on the curated set of core single nucleotide polymorphisms (SNPs) inferred to be outside of recombinant segments by the program Gubbins (Croucher et al., 2015). Colored branches and blocks represent the host species of origin for the various isolates. The blue star indicates the only isolate of the 45 tested from Banks and Victoria Islands that falls outside of the clonal lineage (from the pre-scapular lymph node of MX10-3). Brackets indicate cases of a clone shared between animals. Greek letters (α, β, δ, λ) represent cases of polyclonal infection within individual caribou, with isolates from a single individual shown by the same symbol. The red asterisk indicates the lineage within Clade 3 that contains only isolates from northern wildlife. (C) This ML tree generated using reference-based SNP calls shows the relationship among the 44 isolates within the clonal lineage from Banks and Victoria Island muskoxen (shown in greater detail in Figure 3). The asterisk denotes the dominant branch within this group of isolates. VI, Victoria Island; BC, British Columbia; AB, Alberta.