Figure 1.
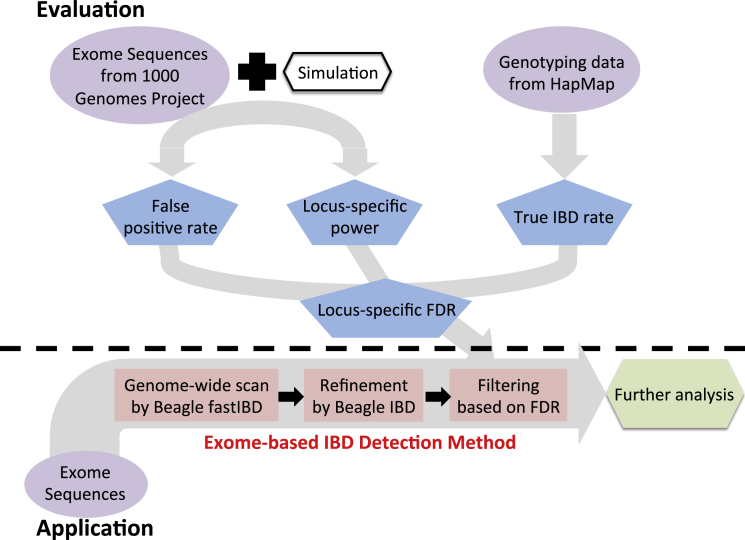
Evaluation and Implementation of ExIBD
In the first step (identification), candidate IBD segments are detected for the whole genome using Beagle fastIBD with fastibdthreshold = 10−10. In the second step (refinement), endpoints of candidate IBD segments are refined with Beagle IBD with ibd2nonibd = 0.01 and nonibd2ibd = 0.0001. Finally, in the third step (filtering), IBD segments from the call set that are likely false positives using the locus-specific FDR are removed. The locus-specific FDR is estimated from the locus-specific power, false-positive rate, and the true IBD rate in the population.