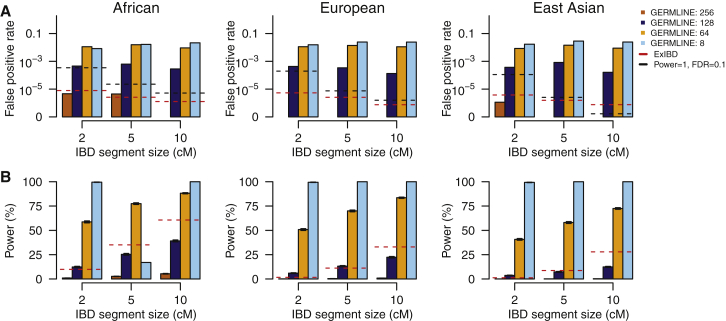
Figure 5.
Performance of GERMLINE under Different Parameter Settings
(A) The false-positive rate to detect IBD segments with different size intervals (e.g., 1.5–2.5 cM, 4.5–5.5 cM, and 9.5–10.5 cM).
(B) The average locus-specific power (with 95% confidence interval) to detect IBD segments with size of 2 cM, 5 cM, and 10 cM.
GERMLINE ran by setting the size of slice as 256, 128, 64, and 8, the maximum number of mismatching homozygous markers for a slice as 2, and turn-off of haplotype extension. See Figures S5 and S6 for more results. For comparison, the performance of ExIBD under the default setting (fastibdthreshold = 10−10, ibd2nonibd = 0.01, and nonibd2ibd = 0.0001) was shown in red line. A critical line, any false-positive rates above which can not control FDR < 0.1 even assuming the detection power is as high as 100%, was shown in black line.