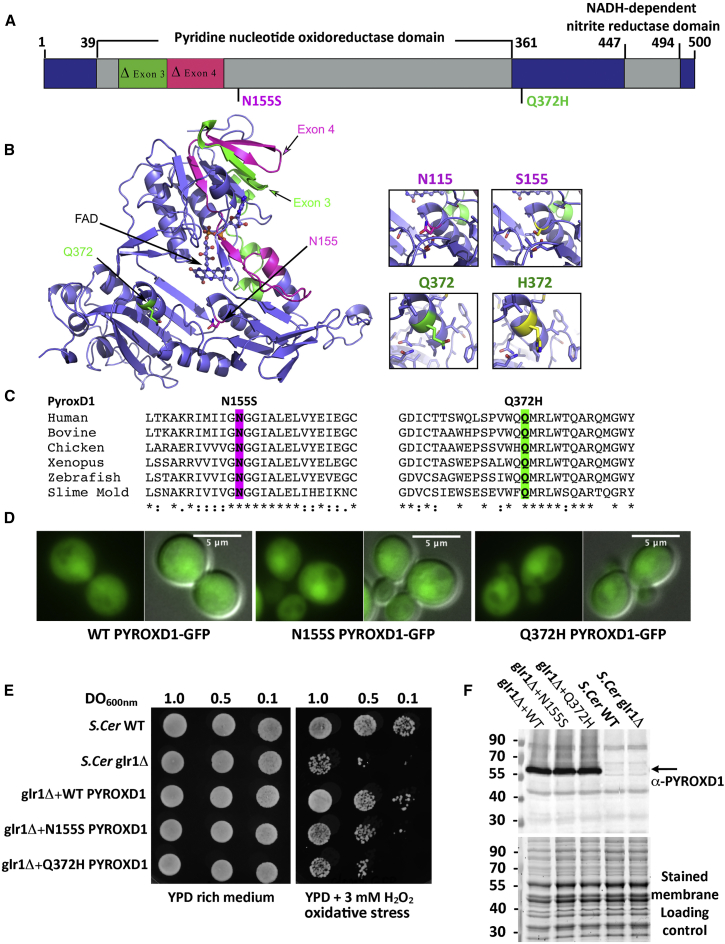
Figure 3.
PYROXD1 Is an Oxidoreductase
(A) A schematic of PYROXD1 with functional domains and identified missense variants created using DOG 2.0.35 Family A, Δexon3 and p.Gln372His (Q372H, green); families B and D, p.Asn155Ser (N155S); family C, Δexon4 and p.Asn155Ser (N155S, red).
(B) PYROXD1 homology model derived from eight homologous crystal structures (see Subjects and Methods). A co-ordinated FAD co-factor and the position of each identified variant on the crystal structure of PYROXD1 are highlighted in green (family A, Δexon3 and Q372H) and pink (family C: Δexon4 and N155S) as in (A).
(C) The identified missense variants p.Asn155Ser and p.Gln372His are evolutionarily conserved to primitive eukaryotes (Uniprot identifiers): human (Q8WU10), bovine (A7YVH9), chicken (F1NPI8), Xenopus (B1WAU8), Danio rerio (Q6PBT5), Dictyostelium (Q54H36).
(D) Living wild-type (WT, BY4742) yeast cells expressing human PYROXD1-GFP, PYROXD1-N155S-GFP, or PYROXD1-Q372H-GFP were observed by fluorescence microscopy with GFP filters and DIC optics. The merge represents the merge between the GFP and DIC images.
(E) Cultures of non-transformed wild-type (WT) or glr1Δ yeast, or glr1Δ yeast transformed with expression plasmids bearing wild-type, N155S-, or Q372H-PYROXD1 were spotted at the indicated concentration (OD600nm) on rich (YPD) or on solid medium containing 3 mM H2O2. Plates were incubated at 30°C and observed after 48 hr.
(F) Western blot of non-transformed wild-type (WT) and glr1Δ, as well as glr1Δ yeast transformed with PYROXD1 expression vectors. The black arrow indicates PYROXD1 and the lower panel shows the protein-stained membrane used as loading control.