Fig. 4.
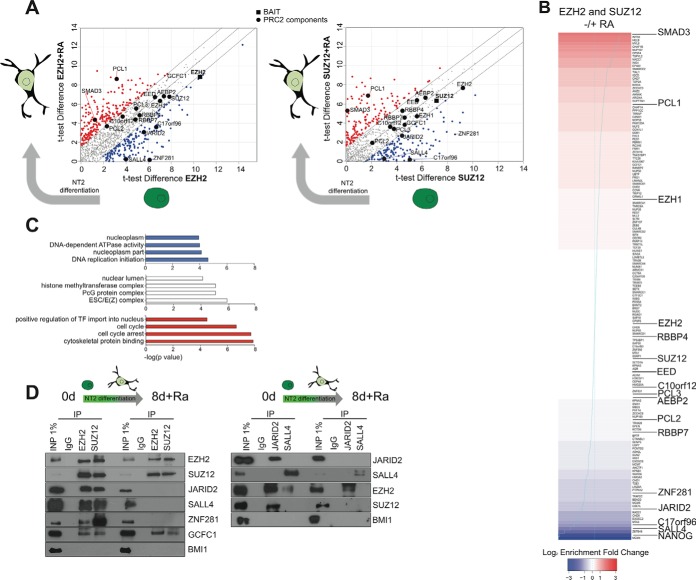
Dynamic PRC2 interactors enriched in alternative cell states. A, t test scores obtained in the undifferentiated and differentiated cell experiments were plotted against each other for EZH2 and SUZZ12, highlighting those proteins favoring interaction with PRC2 in one or other cell state. B, The proteins were ranked according to their fold-change (calculated using the iBAQ values normalized to the immunoprecipitated protein) for combined EZH2 and SUZ12 experiments) favoring interaction in undifferentiated (blue) or differentiated (red) cells. C, Enrichment of Gene Ontology annotations for the proteins favoring static or dynamic interactions. D, Co-immunoprecipitation analysis of candidate static (EZH2, SUZ12, GCFC1) and dynamic (JARID2, SALL4) interactions in both cell states. The PRC1 component BMI1 serves as a negative control.