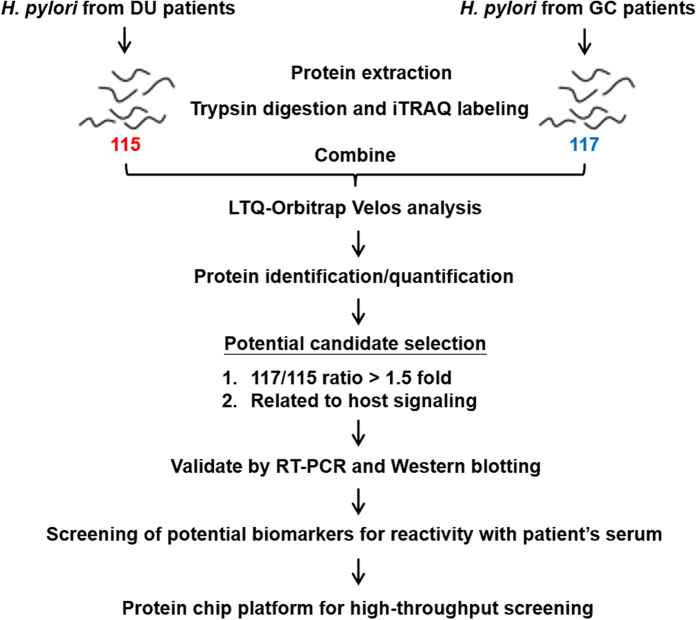
Figure 1. Flow diagram for the identification of differentially expressed virulence factors in different H. pylori strains.
Proteins from pooled H. pylori strains from DU patients or GC patients (n = 5 per group) were digested and labeled separately with different iTRAQ reagents in three independent repeats, then the labeled peptides were combined and separated on a LTQ-Orbitrap Velos hybrid mass spectrometer. Increased expression (>1.5-fold) in GC-derived H. pylori strains than in DU-derived H. pylori strains and proteins related to host signaling by KEGG pathway mapping were used as criteria for selection of candidates, and the candidates were validated by RT-PCR and Western blotting. The selected candidates were then verified by reaction with patients’ serum samples, and a GC-related protein microarray platform was established to perform rapid diagnosis testing.