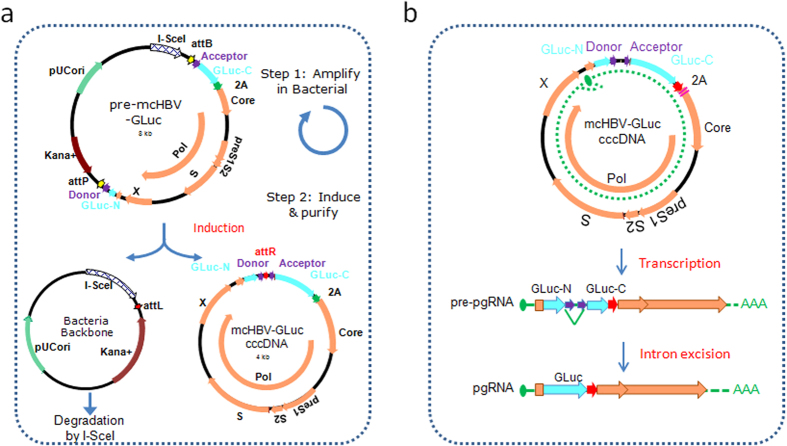
Figure 1. Generation of a novel functional mcHBV-GLuc cccDNA model.
(a) Strategy to generate mcHBV-GLuc cccDNA in E. coli. The pre-mcHBV-GLuc in the plasmid backbone can be amplified in E. coli. After recombinase induction, pre-mcHBV-GLuc is subjected to attP and attB recombination and produces two DNA circles. One with the bacteria backbone (left)is degraded by the I-SceI, while the other (right) mcHBV-GLuc cccDNA is maintained. pUCori: plasmid replication origin; Kana + : kanamycin resistance gene coding sequence; I-SceI: tandem I-SceI cutting sites; attB and attP: recombinase recognizing sites; attL and attR: sequences generated by attB and attP recombination; Donnor and Acceptor: Splicing donor and acceptor sequences; GLuc-N and GLuc-C: N and C terminal of GLuc coding sequences; 2A, Core, Pol, PreS1/S2/S, X are as in Fig. 1a. (b) GLuc expression from mcHBV-GLuc cccDNA. GLuc gene (blue) is split into GLuc-N and GLuc-C by an intron (purple, with splicing donor and acceptor sequence). Pre-pregenomic RNA (Pre-pgRNA) transcript is generated from mcHBV-GLuc cccDNA. The intron is then spliced out and GLuc-N and GLuc-C are ligated to form the functional Gluc mRNA.