Summary
Increased tolerance of crops to low oxygen (hypoxia) during flooding is a key target for food security. In Arabidopsis thaliana (L.) Heynh., the N‐end rule pathway of targeted proteolysis controls plant responses to hypoxia by regulating the stability of group VII ethylene response factor (ERFVII) transcription factors, controlled by the oxidation status of amino terminal (Nt)‐cysteine (Cys). Here, we show that the barley (Hordeum vulgare L.) ERFVII BERF1 is a substrate of the N‐end rule pathway in vitro. Furthermore, we show that Nt‐Cys acts as a sensor for hypoxia in vivo, as the stability of the oxygen‐sensor reporter protein MCGGAIL‐GUS increased in waterlogged transgenic plants. Transgenic RNAi barley plants, with reduced expression of the N‐end rule pathway N‐recognin E3 ligase PROTEOLYSIS6 (HvPRT6), showed increased expression of hypoxia‐associated genes and altered seed germination phenotypes. In addition, in response to waterlogging, transgenic plants showed sustained biomass, enhanced yield, retention of chlorophyll, and enhanced induction of hypoxia‐related genes. HvPRT6 RNAi plants also showed reduced chlorophyll degradation in response to continued darkness, often associated with waterlogged conditions. Barley Targeting Induced Local Lesions IN Genomes (TILLING) lines, containing mutant alleles of HvPRT6, also showed increased expression of hypoxia‐related genes and phenotypes similar to RNAi lines. We conclude that the N‐end rule pathway represents an important target for plant breeding to enhance tolerance to waterlogging in barley and other cereals.
Keywords: N‐end rule, waterlogging, targeted proteolysis, ERFVIIs, PRT6
Introduction
Oxygen is required in plants for energy production through respiration, and low‐oxygen conditions (hypoxia) lead to dramatically changed metabolism to provide alternative sources of ATP (Banti et al., 2013). Hypoxic conditions can occur through changes in the environment surrounding the plant (e.g. flooding), through physical barriers imposed by plant anatomy or during developmental processes with high energy demands (Bailey‐Serres and Voesenek, 2008; Bailey‐Serres et al., 2012; Banti et al., 2013; Vashisht et al., 2011). During flooding events, plant roots can become waterlogged and aerial parts may become submerged in turbid water, leading to reduced capacity for respiration and photosynthesis. Prolonged exposure to hypoxic conditions leads ultimately to cell death.
There are an increasing number of flooding events worldwide, possibly as a result of climate change, and therefore, there is an imperative to provide crop types with increased capacity to tolerate low‐oxygen conditions (Bailey‐Serres et al., 2012; Voesenek and Bailey‐Serres, 2013). Whereas aquatic flowering plants can tolerate submergence, few cultivated crop species show low‐oxygen tolerance mechanisms. Semi‐aquatic rice (Oryza sativa) shows two opposite strategies associated with flooding evasion, quiescence or escape (Bailey‐Serres and Voesenek, 2008; Bailey‐Serres et al., 2012). A quiescence response is controlled by the SUBMERGENCE1A (SUB1A) locus and relies on a suppression of growth and induction of fermentative metabolism until the stress is removed (Bailey‐Serres et al., 2009). The SUB1A locus is not present in standard breeding lines, but was originally identified in submergence‐tolerant lowland rice land races from the Indian subcontinent, and is now used extensively in breeding programmes (Bailey‐Serres et al., 2009; Xu et al., 2006). In contrast to SUB1A‐induced quiescence, deep‐water rice varieties may escape flooding through increased internode elongation, controlled by the SNORKEL1(SK1) and SK2 loci (Hattori et al., 2009). In both cases, the genes controlling these responses have been cloned and shown to encode representatives of the group VII ethylene response factor (ERF) transcription factor subfamily (Hattori et al., 2009; Xu et al., 2006). Quiescence and escape strategies have also been identified recently in two Rumex species from contrasting hydrological niches (van Veen et al., 2013).
Barley is comparatively more susceptible to waterlogging than other cereals. Flooding is a limiting factor for barley production in regions with high rainfall, and average yield can be reduced by up to 50% as a result of waterlogging (Setter and Waters, 2003). Although there is a low heritability for waterlogging tolerance in barley, resistance to this stress is an important objective of breeding efforts in high‐rainfall areas of the world. Leaf chlorosis is an early symptom of barley waterlogging, associated with reduced chlorophyll content, reduction in photosynthesis and senescence, although a differential response to waterlogging in barley varieties was linked to the pattern of aerenchyma formation (Pang et al., 2004; Zhou, 2011). It has been suggested that selecting genotypes with least reduction in photosynthetic rate or total chlorophyll content may facilitate identification of tolerant types (Pang et al., 2004). Quantitative trait loci (QTL) associated with tolerance to waterlogging in barley have been identified using combinations of leaf chlorosis, plant biomass production and plant death (Li et al., 2008, 2013; Mano and Takeda, 2012; Zhou, 2011; Zhou et al., 2012).
Two independent studies in arabidopsis (Arabidopsis thaliana (L.) Heynh.) demonstrated that the evolutionarily conserved N‐end rule pathway of targeted proteolysis is a key mechanism involved in the low‐oxygen response in plants (Gibbs et al., 2011; Licausi et al., 2011). The N‐end rule pathway relates the fate of a protein with the identity of its N‐terminal (Nt)‐residue (Bachmair et al., 1986; Gibbs et al., 2014a; Varshavsky, 2011). Proteins containing destabilizing Nt residues (N‐degrons) are ubiquitinated by specific E3 ligases (N‐recognins) and targeted for proteosomal degradation (Figure 1a). N‐degrons can be created by proteolytic, enzymatic or chemical modification of the Nt‐residue. In arabidopsis, PROTEOLYSIS6 (PRT6) is the N‐recognin for the arginine (Arg) branch of the N‐end rule pathway (Arg/N‐end rule) (Garzon et al., 2007; Graciet et al., 2009, 2010; Holman et al., 2009). Members of the group VII ERF (ERFVII) transcription factor subfamily were shown to be the substrates for this pathway, acting as oxygen sensors (Gibbs et al., 2011; Licausi et al., 2011). The arabidopsis subfamily includes five members, RELATED TO AP2 (RAP2) 2.12, 2.2, 2.3 AND HYPOXIA RESPONSIVE (HRE) 1 and 2 (Licausi et al., 2010; Nakano et al., 2006), each possessing a characteristic N‐terminal motif MCGGAII/L (Nakano et al., 2006). Constitutive removal of Nt‐methionine (Met) by Met aminopeptidase (MAP) (Frottin et al., 2006; Ross et al., 2005) reveals Nt‐Cys, a tertiary destabilizing residue (Varshavsky, 2011). An exposed N‐terminal Cys residue is vulnerable to oxidation, which permits subsequent arginylation by Arg‐tRNA protein transferases (ATEs; Figure 1a), followed by ubiquitination by N‐recognins recognizing the Arg destabilizing residue. Hence, Nt‐Cys acts as a sensor for oxygen (Hu et al., 2005; Lee et al., 2005) through the Cys‐Arg branch of the N‐end rule pathway. It was also shown that both nitric oxide (NO) and oxygen are required for conversion of Cys to its fully oxidized state (Cys‐sulphonic acid) and degradation of Nt‐Cys‐substrates and that plant‐specific enzymes (plant cysteine oxidases; PCOs) can contribute to this oxidation step using oxygen as a cofactor (Gibbs et al., 2014b; Hu et al., 2005; Weits et al., 2014). The rice SUB1A‐1 protein, though an ERFVII, was shown in vitro not to be a substrate of the N‐end rule pathway, suggesting that manipulation of components of the N‐end rule pathway to stabilize endogenous substrates may lead to enhanced tolerance to hypoxic environmental conditions. Two barley (Hordeum vulgare L.) ERFVIIs have previously been described and characterized; HvRAF was shown to be induced in barley seedlings by various treatments and conferred pathogen resistance and salt tolerance in transgenic arabidopsis (Jung et al., 2007), whereas BERF1 is involved in fine‐tuning of expression of the Barley knox3 (Bkn3) gene by ethylene (Osnato et al., 2010).
Figure 1.
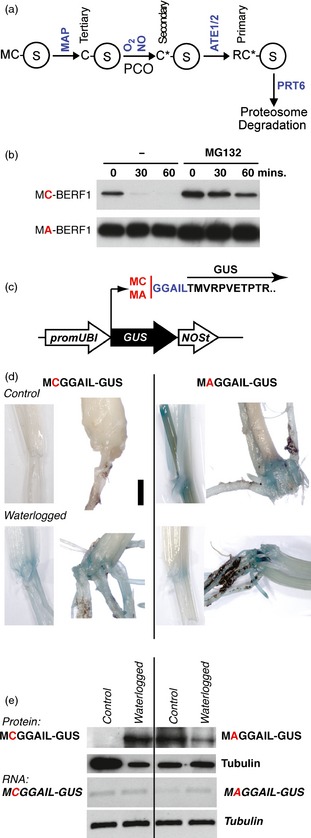
Amino terminal cysteine controls protein stability in barley. (a) The Cys‐Arg/N‐end rule pathway. Amino acids are indicated by single letters; C*, oxidized cysteine; MAP, methionine aminopeptidase; NO, nitric oxide; ATE, arginyl tRNA transferase; PRT6 E3, ligase PROTEOLYSIS6; and PCO, plant cysteine oxidase. Circled S indicates substrate proteins. Tertiary, secondary and primary Nt‐destabilizing residues are indicated. (b) α‐HA Western blot analysis of the in vitro stability of HA‐tagged wild‐type and Ala2 variants of barley ERF1 (BERF1) in the absence or presence of MG132 following treatment with 100 μm cycloheximide at time point 0. Equal loading was confirmed by ponceau staining (data not shown). (c) Diagrammatical representation of the M(C/A)GGAIL‐GUS reporter construct used to analyse substrate stability in vivo in response to waterlogging. (d) Histochemical staining for GUS activity in transgenic barley leaf and root tissue following growth under well‐drained or waterlogged conditions for 15 days. In waterlogged conditions, the analysed tissue was submerged. (e) α‐GUS Western blot analysis of M(C/A)GGAIL‐GUS reporter and tubulin protein stability in response to waterlogging. Expression of RNA via semi‐quantitative rtPCR showing no change in GUS RNA expression in MC‐ or MA‐ constructs in response to waterlogging.
In this study, we demonstrate that barley possesses a functional Cys‐Arg/N‐end rule pathway and that Nt‐Cys, as part of the ERFVII consensus Nt‐sequence, acts as a sensor of waterlogging. We show that reduction in the expression or mutation of the barley N‐recognin E3 ligase HvPRT6 gene leads to an increased tolerance to waterlogging, which is associated with changes in specific flooding‐related traits such as leaf chlorosis, chlorophyll content and biomass. Our work shows that the N‐end rule pathway and its proteolytic substrate proteins represent targets for the manipulation of resistance to waterlogging in barley.
Results and discussion
The barley ERFVII BERF1 is a substrate of the N‐end rule pathway, and amino terminal (Nt‐) Cys acts as a sensor for waterlogging status in barley
The ERFVIIs are characterized by the presence of the Nt‐conserved sequence MCGGAI(I/L) (Nakano et al., 2006). This sequence acts to sense oxygen and nitric oxide (NO) via the N‐end rule pathway of targeted proteolysis through the Nt‐Cys that is exposed following the action of MAPs (Figure 1a) (Gibbs et al., 2011, 2014b; Hu et al., 2005; Licausi et al., 2011). We searched the barley proteome (Mayer et al., 2012) initially to identify AP2‐domain proteins initiating MC‐ and then to define those with the consensus MCGGAI(I/L). There are 13 AP2‐domain‐containing proteins initiating MC‐ but only 7 sharing significant sequence similarity to the consensus Nt‐sequence of ERFVIIs (Figure S1, Table S1). Similarly, in rice, several ERFVIIs do not contain the full Nt‐consensus sequence (Gibbs et al., 2011). It is possible to distinguish subgroups with similarity to arabidopsis and wheat ERFVIIs (Figure S1b). We investigated whether barley ERFVIIs might be substrates of the N‐end rule pathway using an in vitro heterologous rabbit reticulocyte lysate assay (Gibbs et al., 2011; Lee et al., 2005), as components of the N‐end rule pathway are evolutionarily conserved (including enzyme activities of MAP, ATEs and UBR1 orthologue PRT6) (Graciet et al., 2010). We focused on the barley ERFVII family member BERF1, as it is most closely related in sequence to RAP2.12 in arabidopsis (Figure S1b), proposed to be the major oxygen sensing member of the subfamily (Licausi et al., 2011) and TaERF1 from wheat (Xu et al., 2007). We expressed C‐terminally haemagglutinin (HA)‐tagged BERF1 in this system and analysed the relative stability of native (Cys2) and mutated (Ala2) versions of the proteins in the presence and absence of the proteasome inhibitor MG132 following treatment with cycloheximide to block translation (Figure 1b). Whereas Cys2 protein was rapidly degraded in this system, and stabilized by MG132, conversion of Cys2 to Ala (a stabilizing residue; (Varshavsky, 2011)) greatly enhanced stability, indicating that BERF1 may be a substrate of the N‐end rule pathway in vivo.
To demonstrate that Nt‐Cys, as part of the barley ERFVII Nt‐consensus, can act as a sensor for hypoxia in vivo in transgenic barley plants, we analysed the stability of a modified β‐glucuronidase (GUS) reporter protein initiating with the consensus first 7 amino acids of the ERFVIIs, MCGGAIL‐ (or a mutated Ala2 version MAGGAIL‐) (Figure 1c). This reporter was previously used to demonstrate that Nt‐Cys senses NO in barley (Gibbs et al., 2014b). In this construct, transgene RNA expression is driven constitutively by the maize ubiquitin promoter (Bartlett et al., 2008). Under well‐drained normal growth conditions, no GUS staining could be observed in plants expressing the WT MCGGAIL‐GUS construction, whereas activity of the mutated Ala2 version (MAGGAIL‐GUS) could be visualized in the hypocotyl and primary roots. To impose waterlogging treatments, pots with seedlings (3–4 well‐developed leaves) were placed for 2 weeks in a plastic container filled with water. During the experiment, the level of water was maintained to ensure that the hypocotyl was covered. As a result of waterlogging, GUS activity could also be visualized in plants containing MCGGAIL‐GUS (Figure 1d) and Western blot analysis showed stabilization of GUS protein, which occurred in the absence of any changes in transgene mRNA levels (Figure 1e). These results demonstrate that Nt‐Cys, as part of the ERFVII consensus MCGGAIL, has the capacity to act as a sensor of waterlogging status. In arabidopsis, it was shown that HRE2 and RAP2.3 protein stability was enhanced by both hypoxia and submergence (Gibbs et al., 2011, 2014b), although no direct effect of root waterlogging on substrate stability has been shown. In conjunction with in vitro results using BERF1, our results suggest that barley ERFVIIs act as regulators of the barley response to waterlogging through their function as sensors of hypoxia via Nt‐Cys.
Reducing expression of the barley N‐recognin E3 ligase HvPROTEOLYSIS6 (HvPRT6) increases expression of hypoxia‐related genes
The single‐copy gene PROTEOLYSIS6 (PRT6) encodes the N‐recognin for the Arg branch of the N‐end rule pathway (Figure 1a) in arabidopsis (Garzon et al., 2007). We analysed the copy number of PRT6 orthologues in other species using proteomes derived from sequenced plant genomes, including rice (Yu et al., 2002) (LOC_Os01g05500), Brachypodium distachyon (Vogel et al., 2010) (Bradi2g03180) and barley (Mayer et al., 2012) (MLOC_47469.1). In all cases, only a single protein was identified (Figure S2), and important domains for function, including the UBR box, RING finger domain and auto‐inhibitory domain, were found to be highly conserved across all species. Analysis of RNA levels for the barley PRT6 orthologue (HvPRT6) indicates constitutive expression during growth and development (Figure S3). To test the role of HvPRT6 in regulating barley response to waterlogging, an RNAi approach was used. An RNAi construct was designed targeting a 600‐bp exon sequence corresponding to the region of the protein between the UBR box and RING domain features (Figure S2). Several lines derived from independent transformation events were generated, containing different copy numbers at single insertion sites for the RNAi construct (Table S2). Homozygous transgenic lines and associated null lines (nontransgenic but derived from the same transformation event) were initially analysed to observe if the introduced construct altered expression of the endogenous HvPRT6 gene. In all cases, a large reduction in expression of HvPRT6 RNA was observed (Figure 2). Genetic removal of the function of N‐end rule components in arabidopsis, including ATE and PRT6 (Figure 1a), leads to an increase in expression of genes associated with the core hypoxia response, including ALCOHOL DEHYDROGENASE1 (ADH1), due to constitutive stabilization of ERFVIIs (Gibbs et al., 2011; Licausi et al., 2011; Mustroph et al., 2009). Analysis of leaf material of barley RNAi lines by quantitative (q) RT‐PCR revealed an increase in expression of the barley homologues of ADH (HvADH1), HAEMOGLOBIN (HvHB) and PYRUVATE DECARBOXYLASE1 (HvPDC1) compared to untransformed controls. This indicates that reduction in HvPRT6 activity via RNAi silencing leads to an increased hypoxia‐associated gene expression. These increases were not as dramatic as seen in the arabidopsis prt6 null mutant, which is likely due to the RNAi lines not completely abolishing expression of this gene.
Figure 2.
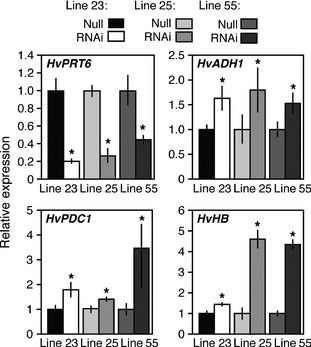
qRT‐PCR analysis of the influence of the HvPRT6 RNAi construct on gene expression in independent barley homozygous RNAi lines compared to un‐transformed null segregant controls. Relative expression of HvPRT6, HvADH1, HvHB, HvPDC1. Transcript levels are shown relative to respective null segregants. In each case, error bars represent standard deviation of the mean. *P < 0.05.
Reduced HvPRT6 expression influences mature seed phenotypes
The N‐end rule pathway controls many plant developmental and environmental responses (Gibbs et al., 2011, 2014a,b; Graciet et al., 2009; Holman et al., 2009; Licausi et al., 2011). In particular, it was shown that the pathway promotes seed germination and reduced abscisic acid (ABA) sensitivity by enhancing the degradation of ERFVIIs in response to NO (Gibbs et al., 2014b; Holman et al., 2009). Whereas in arabidopsis, light stimulates germination during imbibition, in barley, germination and removal of dormancy are enhanced in the dark (Gubler et al., 2008). We found that after‐ripened barley seeds completed germination rapidly in the dark, with germination being slightly delayed in the RNAi lines (Figure 3a). However, RNAi lines showed a severe reduction in germination in comparison with null controls in the light. These data strongly indicate that the N‐end rule pathway controls germination in cereals, and reduced germination in white light in the RNAi lines may result from increased stability of ERFVII substrates.
Figure 3.
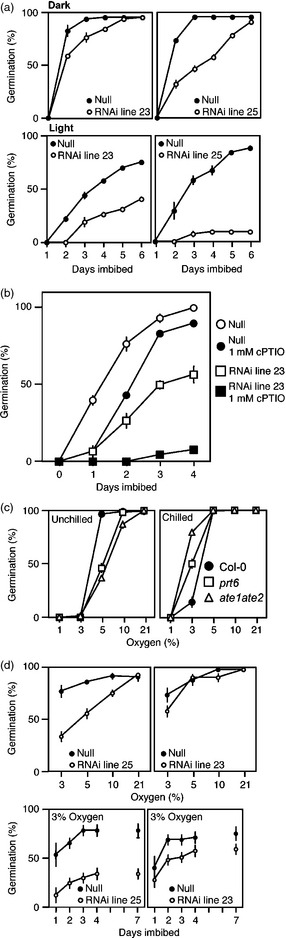
Reduced HvPRT6 expression alters barley seed germination phenotypes. (a) Germination of two independent barley homozygous RNAi lines compared to un‐transformed null segregants under dark (top) or white light (bottom) conditions. (b) The influence of the NO scavenger cPTIO on germination in the light. (c) Germination of un‐chilled and chilled arabidopsis WT (Col‐0), prt6 and ate1ate2 seeds at 22 °C in continuous light and under reduced oxygen availability. (d) Germination of barley seeds at 21 °C under reduced oxygen availability following moist chilling. Final germination percentage at each oxygen concentration (top) and germination rate over 7 days at 3% oxygen (bottom) is shown. In each case, error bars represent standard deviation of the mean of three independent experiments.
To link the Cys‐Arg branch of the N‐end rule pathway to the regulation of germination in barley, we analysed the influence on RNAi lines of the cell‐permeant NO scavenger 2‐(4‐carboxyphenyl)‐4,4,5,5‐tetramethylimidazoline‐1‐oxyl‐3‐oxide (cPTIO). Previously, we showed that the MCGGAIL‐GUS reporter was stabilized in barley embryos treated with cPTIO, suggesting that substrates of the Cys‐Arg/N‐end rule pathway act as sensors for NO in barley (Gibbs et al., 2014b). In addition, it was previously shown that NO can break dormancy in both barley and arabidopsis (Bethke et al., 2004), and arabidopsis dormancy is regulated specifically by ERFVII stability in response to NO (Gibbs et al., 2014b). We found that null segregant barley seeds showed a small reduction in germination speed in the presence of cPTIO in the light, whereas germination of RNAi seeds was almost completely inhibited (Figure 3b). These data indicate that in barley seeds, the N‐end rule pathway controls substrate stability and germination potential in a similar way to arabidopsis (Gibbs et al., 2014b; Holman et al., 2009). It has been suggested that the barley coleorhiza (an embryo‐derived tissue that covers the emerging root) may play a similar role to the arabidopsis endosperm (Barrero et al., 2009), implying that by analogy, the N‐end rule pathway may regulate barley germination in response to NO via crosstalk with ABA signalling in this tissue.
Arabidopsis prt6 and ate1ate2 mutant seeds that were moist chilled (to remove dormancy) showed enhanced germination under low oxygen compared to wild‐type (WT; accession Col‐0) seeds, which is presumed to result from constitutive expression of hypoxia‐survival genes in the mutants (Gibbs et al., 2011). However, we found that the response of un‐chilled arabidopsis seeds to hypoxia was very different. Whereas WT seeds showed almost complete germination even in 5% oxygen, seeds of prt6 and ate1ate2 showed a reduced ability to germinate (Figure 3c). Moist chilling of arabidopsis seeds has previously been shown to strongly repress ABA sensitivity and enhance GA sensitivity (Holman et al., 2009; Ogawa et al., 2003; Yamauchi et al., 2004). In barley, germination under hypoxia is strongly repressed by ABA and the surrounding hull (Benech‐Arnold et al., 2006; Bradford et al., 2008; Hoang et al., 2013; Mendiondo et al., 2010). We tested the germination sensitivity of two barley RNAi lines at different oxygen levels following moist chilling and found that in comparison with null segregants, the final germination levels and speed of germination were reduced at the lowest oxygen level (Figure 3d). These results, similar to what is observed in nonchilled arabidopsis N‐end rule mutants, indicate that stabilization of N‐end rule substrates reduces germination capacity under hypoxia, which may result from increased ABA sensitivity.
Reduced HvPRT6 expression increases tolerance to waterlogging
Constitutive overexpression of native arabidopsis RAP2.12 (35S:RAP2.12) enhances survival in response to submergence and increases hypoxia‐related gene expression (Licausi et al., 2011). However, it was also shown in the same report that plant tolerance to flooding decreased in both prt6 and ate1ate2 mutants. This is in contrast to what is observed in rice lines containing the SUB1A locus, which have dramatically enhanced submergence tolerance (Bailey‐Serres et al., 2009). To determine the influence of reduced HvPRT6 expression on tolerance to flooding, we examined the growth and physiological responses of barley HvPRT6 RNAi lines to waterlogging. Following waterlogging treatment (20 days), several obvious differences in growth and development between nontransgenic and RNAi plants were observed. In comparison with null segregants, RNAi lines showed continued growth and maintenance of green leaves (Figure 4a). To quantify the responses to waterlogging, we analysed hypoxia‐related gene expression, chlorophyll levels, biomass and final yield. The expression of hypoxia‐related genes HvADH1, HvHB and HvPDC1 in response to waterlogging was greater in RNAi lines than in null segregant controls (Figure 4b). One obvious difference in response to waterlogging was the retention of green coloration of transgenic leaves compared to controls, possibly as a result of enhanced maintenance of leaf chlorophyll content in response to the treatment. To investigate this, chlorophyll was extracted and quantified from leaves of null segregant and RNAi plants following waterlogging (Figure 4c). This analysis revealed a clear retention of chlorophyll in transgenic leaves compared to controls, suggesting that active photosynthesis continued in these lines despite waterlogging and also that leaf senescence was reduced. Both of these traits have previously been identified as hallmarks of waterlogging tolerance in barley, and we hypothesized that these factors may contribute to the relative enhanced growth of transgenics in excess water conditions. In agreement with this, analysis of total above‐ground biomass in response to waterlogging demonstrated that RNAi lines were relatively insensitive to the waterlogging treatment (Figure 4d), showing significantly less reduction in total plant weight in response to waterlogging compared to null segregants. As a consequence of this, transgenic lines also showed sustained yield under waterlogging conditions (Figure 4e). Collectively, our physiological analyses indicate that reducing HvPRT6 levels has a positive effect on general growth and survival during waterlogging.
Figure 4.
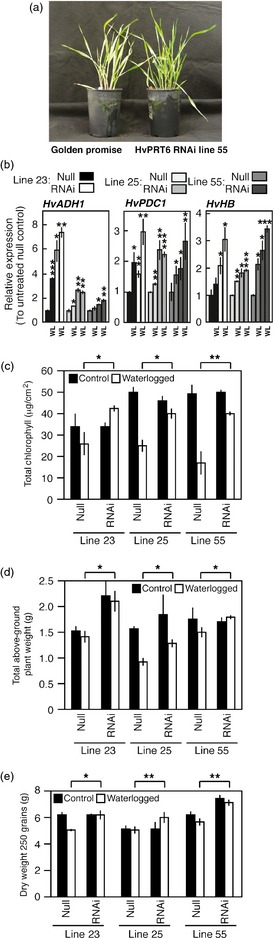
Reduced HvPRT6 expression alters barley response to waterlogging. (a) Photograph of 25‐day‐old plants following 20 days of waterlogging, showing enhanced green leaf material in RNAi line 55 compared to WT. (b) Relative expression of hypoxia‐related genes measured by qRT‐PCR from healthy leaves of null and RNAi lines grown under well‐drained or waterlogged (WL) conditions. Transcript levels and significance are shown relative to well‐drained null segregant controls. (c) Total chlorophyll derived from leaves of plants grown under well‐drained or waterlogged conditions. (d) Total above‐ground plant weight following growth under well‐drained (control) or waterlogged conditions. (e) Total yield (dry weight of 250 grains) following growth under well‐drained (control) or waterlogged conditions. In each case, error bars represent standard deviation of the mean. *P < 0.05, **P < 0.01, ***P < 0.001.
Reduced HvPRT6 expression enhances the maintenance of chlorophyll in leaves in continued darkness
One symptom of plants exposed to prolonged foliar submergence is leaf senescence and chlorophyll degradation (Fukao et al., 2012; Vashisht et al., 2011). The effect of the HvPRT6 RNAi construct on dark‐induced senescence was analysed using leaf segments incubated on one‐half‐strength Murashige and Skoog (MS) medium in darkness. Following 6 days of dark treatment, nontransgenic leaf segments appeared less green than those from RNAi lines (Figure 5a). To quantify this observation, we developed an image analysis approach using a transformation of images of the leaf segments from RGB colour space (red, green and blue) to HSV colour space (hue, saturation and value). Red, green and blue contributions were quantified, and from this, we calculated the hue of individual leaf patches (Figures S4, S5 and S6). As a result, we obtained a good measure of how green or yellow a leaf segment appears, allowing a quantification of colour change (Figure 5b). Using this procedure, it was possible to discriminate with statistical significance between null segregant and transgenic lines, showing that the colour of transgenic leaf sections did not alter in response to treatment as much as nontransgenic leaf sections. This implies a delay in leaf senescence in transgenics, similar to that previously observed in SUB1A in rice (Fukao et al., 2006, 2012). This suggests that stabilization of substrates in HvPRT6 RNAi lines leads to enhanced tolerance to extended darkness, which may occur through repression of pathways activating leaf senescence and chlorophyll breakdown.
Figure 5.
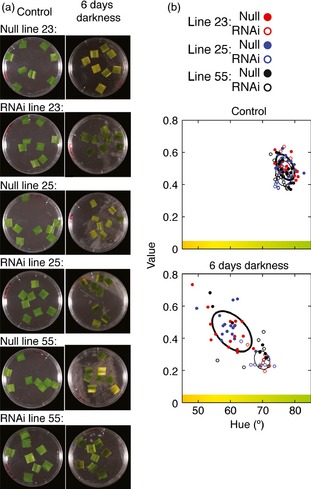
Reduced HvPRT6 expression delays leaf senescence. (a) Photographs of leaf sections from RNAi lines and nontransgenic controls following treatment with darkness on liquid media for 6 days. (b) Quantification of colour change in leaf sections following transformation of images of leaf segments from RGB to HSV colour space. The 50% confidence regions are shown as ellipses (thick for null lines and thin for RNAi lines).
Identification and analysis of Targeting Induced Local Lesions In Genomes (TILLING) lines containing mutant alleles of HvPRT6
To further investigate the role of HvPRT6 in barley waterlogging responses, we screened a‐mutagenized M2 population of barley variety Sebastian using the TILLING method (Kurowska et al., 2011; Weil, 2009) and identified two lines containing mis‐sense mutations in HvPRT6, prt6i (resulting in a proline to serine substitution; P1583S) and prt6k (resulting in an alanine to threonine substitution; A1543T) located between the Cys‐/His‐rich Ring domain and the C‐terminal auto‐inhibitory domain (Figure S2, Figure 6a). The prt6k mutation could be interpreted as conservative, as the arabidopsis PRT6 protein also contains threonine at this position, whereas the prt6i substitution changes a proline conserved in flowering plant PRT6 proteins. In both cases, mutant alleles resulted in decreased expression of HvPRT6 RNA (Figure S7). Hypoxia‐related phenotypes and gene expression were analysed in the TILLING lines homozygous for prt6 mutations and compared to WT (variety Sebastian). Growth was less affected by waterlogging in both lines compared to the WT (Figure 6b), chlorophyll levels were higher in TILLING lines following waterlogging (Figure 6c), and the expression of hypoxia‐related genes was increased in mutant lines relative to WT under both normal and waterlogged conditions (Figure 6d). Furthermore, dark‐induced leaf senescence was also delayed in the TILLING lines, as observed for RNAi lines (Figure 6e,f). Collectively, these data suggest that both TILLING lines have reduced HvPRT6 protein activity and corroborate our RNAi data to suggest that the N‐end rule pathway is a key regulator of waterlogging responses in barley. Further development of HvPRT6 TILLING lines and analysis under field growth conditions may provide useful resources for breeders.
Figure 6.
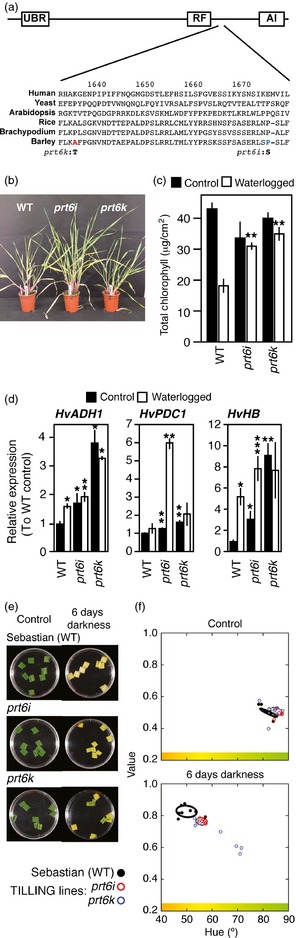
Phenotypes of two barley TILLING lines containing mutations in PRT6. (a) Positions of amino acid sequence changes in prt6i and prt6k TILLING lines. (b) Photograph of 20‐day‐old plants following 20 days of waterlogging, showing enhanced growth of TILLING lines compared to WT (Sebastian). (c) Total chlorophyll derived from leaves of plants grown under well‐drained or waterlogged conditions. (d) Expression of hypoxia‐related genes measured by qRT‐PCR from leaves of plants grown under well‐drained (control) or waterlogged conditions. (e) Photographs of leaf sections from RNAi lines and nontransgenic controls following treatment with darkness in 1/2MS liquid media for 6 days. (f) Quantification of colour change in leaf sections following transformation of images of leaf segments from RGB to HSV colour space. The 50% confidence regions are shown as ellipses (thick for Sebastian and thin for TILLING lines). *P < 0.05, **P < 0.01, ***P < 0.001.
Our results indicate that manipulation of the N‐end rule pathway may provide approaches to increase tolerance to waterlogging, a stress that significantly affects barley productivity in areas with high rainfall. Collectively, our findings suggest that the ERFVIIs are N‐end rule substrates in barley that, alongside their known role in arabidopsis, implies that they also function as homeostatic sensors of hypoxia via the N‐end rule pathway in cereals. Transgenic HvPRT6 RNAi lines had higher levels of anaerobic response gene expression, which translated into an increased tolerance to waterlogging, as indicated by stabilized growth and yield, greater chlorophyll retention and delayed dark‐induced leaf senescence under stress conditions relative to WT plants. Future examination of these lines should focus on analysing whether reducing N‐end rule activity positively or negatively affects responses to other stresses, such as drought (Fukao et al., 2011). Future research efforts will focus on targeted stabilization of individual barley N‐end rule substrates and assessment of their role in mediating the response to waterlogging in vivo in this important crop, and the development of nontransgenic TILLING alleles of HvPRT6 to provide resources for breeders.
Experimental procedures
Plant material and growth conditions
The barley varieties Golden Promise (transformation) and Sebastian (TILLING) were used for all experiments. Unless otherwise stated, plants were grown under controlled conditions (15 °C/12 °C; 16‐h photoperiod; 80% RH, 500 μmol/m2/s metal halide lamps (HQI) supplemented with tungsten bulbs). Transgenic barley plants were generated using Agrobacterium‐mediated transformation of immature embryos from the variety Golden Promise (Bartlett et al., 2008). Regenerated transgenic plants were grown under the same conditions. Copy number and zygosity determination were carried out by IDna GENETICS (Norwich, UK). Waterlogging treatments were imposed as follows: well‐drained and waterlogging treatments were applied from developmental stage leaf 7 to leaf 10 (L7‐10). The leaf number at which waterlogging was applied was always measured on the main stem. Waterlogging duration was 15 or 20 days as indicated (de San Celedonio et al., 2014). Crop phenology was determined following the decimal code (Zadoks et al., 1974). To observe the effect of darkness on leaf senescence, fully expanded uppermost leaves were cut into 1‐cm2 sections and floated on one‐half‐strength MS liquid medium in the dark at 22 °C for up to 6 d in sealed Petri dishes (Fukao et al., 2012). Chlorophyll content was determined in triplicate samples by extraction and assay in 80% acetone (Porra et al., 1989). Histochemical staining for GUS activity was carried out as previously described (Gibbs et al., 2014b). Arabidopsis accessions and mutants were the same as those described previously (Gibbs et al., 2011, 2014b).
Sequence identification and analysis
Basic local alignment search tool (BLAST) (Altschul et al., 1990) analysis was carried out to identify the barley orthologue of PRT6. CLUSTALW as part of the MacVector program (MacVector, Cambridge, UK) was used for multiple sequence alignments and to identify barley ERFVIIs from the predicted proteome derived from the barley genome sequence (Mayer et al., 2012). For RNAi silencing, a 530‐bp HvPRT6 cDNA fragment (amplified using primers HvPRT6F and HvPRT6R; Table S3) was cloned into the pCR8GW donor vector (Invitrogen, Paisley, UK) and then into the RNAi destination vector pBract207 (Bartlett et al., 2008).
RNA expression analysis
Total RNA was isolated from young leaf or root tissue as described previously (Mendiondo et al., 2014). Gene‐specific oligonucleotide primers (Table S3) were used in quantitative (q) RT‐PCR with SYBR‐green Sensimix (Bioline, London, UK), in a Roche LightCycler 480 apparatus (Roche/Applied Biosystems, Welwyn Garden City, UK) as specified by the manufacturer. The relative number of copies obtained for each transcript was normalized against HvELF1 and HvTubulin transcript values for each sample as an internal reference (Jarosova and Kundu, 2010; Nicot et al., 2005).
In vitro analysis of protein stability
The entire open reading frame of BERF1 was cloned from cDNA derived from leaf tissue and was directionally cloned into the modified pTNT3xHA vector to produce C‐terminal HA‐tagged fusions either as N‐terminally initiating MC or MA derivatives, and in vitro assays were carried out using rabbit reticulocyte lysate (Gibbs et al., 2011) but with the addition of 100 μm cycloheximide at the 0‐min time point to block mRNA translation.
Western analysis of proteins
Western blots were carried out as previously described (Gibbs et al., 2011, 2014b). Anti‐HA antibodies were obtained from Sigma, and anti‐GUS antibodies were obtained from Invitrogen. Barley leaf samples (100 mg) were ground to powder with a mortar and pestle in liquid nitrogen, and protein extraction was performed (NucleoSpin® RNA/Protein extraction kit, Macherey‐Nagel, Düren‐Germany). Total protein content in the samples was quantified by Bradford protocol with a spectrophotometer (Thermo Scientific Multiskan Ascent, Waltham, MA) against a BSA standard curve. For anti‐GUS Western blots, in each lane, 75 μg of total protein were loaded plus 5 μL of cracking buffer (0.5 m Tris–HCL, pH 6.8; 10% glycerol; 1% SDS; 5% β‐mercaptoethanol; 2 mg bromophenol blue) and water to a final volume of 20 μL. After 0.1% SDS–16.5% PAGE, proteins were electroblotted onto a nitrocellulose membrane. SeeBlue Plus2 Pre‐Stained Standard Marker (Novex, Paisley, UK) was loaded as a reference for protein size. Membranes were reversibly stained with Ponceau S Red to check equal loading and protein integrity.
Seed germination assays
Heads were harvested at maturity, dried for 7 days and threshed by hand to prevent damage to the husk. For each assay, triplicates of thirty grains were placed in Petri dishes containing two layers of Whatman No. 1 filter paper (GE Healthcare Life Sciences, Little Chalfont, UK) and 4 mL sterile water. Dishes were sealed with Micropore tape (3 m) and incubated at 10 °C in the dark or white light. Germinated caryopses, defined by the emergence of coleorhizae beyond the seed coats, were scored every 24 h over 7 days and removed from the dishes. Germination under atmospheres with different controlled oxygen tensions was performed at 21 °C in darkness for barley and 22 °C for arabidopsis as described previously (Côme and Tissaoui, 1968). Gas mixtures containing from 1 to 21% oxygen were obtained through capillary tubes connected to sources of compressed air and nitrogen. The gaseous atmospheres were passed continuously through germination chambers at a constant flow rate (4.0 lh1). Oxygen tensions were measured daily using a Servomex analyser (type 570A; Servomex, Crowborough, UK).
Quantification of leaf section colour
Images of leaf patches were acquired with Canon EOS 5D camera at a resolution of 27 pixels/mm. The size and shape differed between patches. For that reason and because the object could not be readily identified automatically, it was necessary to select sections of each patch for statistical analysis. Manual selection of sections (square or rectangular with an area of approximately 0.3 mm2) of leaves was carried out using the image analysis software Fiji (Schindelin et al., 2012). Subsequently, red, green and blue (RGB) colour channels were analysed in each section with a script written in Fiji's macro programming language (data S1; Figures S4, S5 and S6). Measurements of each region of interest included position within the micrograph, area in square pixels, mean brightness, median brightness and standard deviation of brightness. The mean RGB values of each leaf patch were converted to hue, saturation and value (HSV) using R and RStudio (RDevelopmentCoreTeam, 2012; RStudio, 2012). The formula used to calculate colour hues of yellow leaf patches is 60 degrees × ((G–B)/(R–B) mod 6) (mod refers to the modulo operation, and this formula is a special case for R > G > B and applicable for yellow leaf sections only). For green leaf sections, where G > R > B, the corresponding expression is hue = 60 degrees × ((B–R)/(G–B) + 2). The selection of the correct formula is carried out automatically within the built‐in R function ‘rgb2hsv’. The returned value is an arc length along a regular hexagon with total circumference of one. By multiplying by 360 degrees, an approximation of an angular coordinate is obtained. The structure of the hexagon is still recognizable in the full colour map as shown in Figures S5, S6 and 7, where 0 degrees corresponds to red, 60 to yellow, 120 to green, 180 to turquois, 240 to blue, 300 to magenta and 360 to red. These points correspond to six of the eight corners of the colour cube (taking R, G and B as Cartesian coordinates x, y and z, respectively). The remaining two corners correspond to black and white (which in HSV colour space involves the additional coordinates saturation and value). The 50% confidence regions (ellipses) shown here were obtained with the bivariate boxplot command bv.boxplot of the package asbio for R, implementing a previously described method (Goldberg and Iglewicz, 1992) and a biweight estimator function (Everitt, 2005).
Identification of barley TILLING lines containing mutant alleles of HvPRT6
TILLING lines containing mutant alleles of HvPRT6 were developed from the HorTILLUS (Hordeum vulgare‐TILLING‐University of Silesia) population of spring barley cultivar ‘Sebastian’ created in Department of Genetics, University of Silesia, after double treatment of seeds with sodium azide (NaN3) and N‐nitroso‐N‐methylurea (MNU). Two different treatments were applied to two batches of seeds: 1.5 mM NaN3/3 h–6 h iig–0.75 mm MNU/3 h and 1.5 mm NaN3/3 h–6 h iig–0.5 mm MNU/3 h (iig—interincubation germination period). Mutation screening was performed on DNA isolated from 6,144 M2 HorTILLUS plants. Eleven mutations were found in the HvPRT6 gene, giving a mutation density of 1mut/605 base pairs. Two of these were chosen (prt6i, prt6k) that led to amino acid substitutions, theoretically resulting in an unaffected full‐length protein. For mutation identification, PCR was carried out using genomic DNA from eightfold DNA pools using the primers: forward (F): 5′‐(IRD700)‐ TGTCATGATCGATATTTGTTTTCC‐3′; reverse (R): 5′‐(IRD800)‐ TCGCTTAGTAGCATCCAAAAGA‐3′ that amplify a 695‐bp exon region of HvPRT6 (Figure S2). For heteroduplex formation, samples were incubated at 95 °C for 3 min, and then, slow renaturation was performed in 70 °C per 20 s × 70 cycles (−0.3 °C/cycle). Heteroduplexes were cleaved with Celery Juice Extract as previously described (Till et al., 2006), and DNA fragments were visualized on 6% denaturing polyacrylamide gels. DNA from each individual from positive bulks was then analysed separately to identify single plants carrying mutations within the TILLING region. The DNA sequence of analysed fragments from plants carrying potential mutations in HvPRT6 gene was obtained to confirm the presence of mutation and zygosity state. Seeds from M2 plants carrying mutations in HvPRT6 gene were used for developing homozygous TILLING lines.
Supporting information
Figure S1 A. Alignment of the predicted protein sequences of Group VII ERFs from Arabidopsis thaliana and Hordeum vulgare and wheat TaERF1.
Figure S2 Alignment of the predicted Barley PRT6 sequence with sequelogues from Human (Homo sapiens), yeast (Saccharomyces cerevicieae) and Arabidopsis thaliana.
Figure S3 Expression of the barley PRT6 sequelogue during normal growth and development.
Figure S4 Image analysis of a green and a yellow leaf patch (1 cm2) for quantification of greenness with hue in degrees.
Figure S5 Hue histograms of individual pixels of a green and a yellow leaf patch.
Figure S6 HSV colour space for visualisation of hue and value. Circles are filled with colours specified by their position within the plane of hue and value at maximum saturation of one.
Figure S7 qRT‐PCR analysis of the influence of the HvPRT6 mutant alleles prt6k and prt6i on HvPRT6 gene expression relative to the WT (Sebastian).
Table S1 Genome location of barley ERFVIIs (XLSX file).
Table S2 Transgene copy number in RNAi transgenic lines (XLSX file).
Table S3 List of oligonucleotides used in QrtPCR analyses (XLSX file).
Data S1 ImageJ plug‐in software for analysis of leaf colour (IJM file).
Acknowledgements
GMM, DJG, JFG and MJH were supported by grants from the BBSRC (BB/F006934/1, BB/G010595/1 and BB/K000144/1) including support from SABMiller plc. GMM was supported for part of the work by a Barry Axcell Fellowship. AK was supported by a BBSRC DTP PhD studentship. We thank Zoe Wilson and José Fernández Gómez (University of Nottingham) for guidance with barley transformation, David Marshall and Robbie Waugh (James Hutton Institute) for sharing barely genomic sequences and genome mapping data with us, and Susan Liddell (Sutton Bonington Proteomics Facility).
References
- Altschul, S.F. , Gish, W. , Miller, W. , Myers, E.W. and Lipman, D.J. (1990) Basic local alignment search tool. J. Mol. Biol. 215, 403–410. [DOI] [PubMed] [Google Scholar]
- Bachmair, A. , Finley, D. and Varshavsky, A. (1986) In vivo half‐life of a protein is a function of its amino‐terminal residue. Science, 234, 179–186. [DOI] [PubMed] [Google Scholar]
- Bailey‐Serres, J. and Voesenek, L. (2008) Flooding stress: acclimations and genetic diversity. Annu. Rev. Plant Biol. 59, 313–339. [DOI] [PubMed] [Google Scholar]
- Bailey‐Serres, J. , Fukao, T. , Ronald, P. , Ismail, A. , Heuer, S. and Mackill, D. (2009) Submergence tolerant rice: SUB1's journey from landrace to modern cultivar. Rice, 3, 138–147. [Google Scholar]
- Bailey‐Serres, J. , Fukao, T. , Gibbs, D.J. , Holdsworth, M.J. , Lee, S.C. , Licausi, F. and van Dongen, J.T. (2012) Making sense of low oxygen sensing. Trends Plant Sci. 17, 129–138. [DOI] [PubMed] [Google Scholar]
- Banti, V. , Giuntoli, B. , Gonzali, S. , Loreti, E. , Magneschi, L. , Novi, G. and Perata, P. (2013) Low oxygen response mechanisms in green organisms. Int. J. Mol. Sci. 14, 4734–4761. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barrero, J.M. , Talbot, M.J. , White, R.G. , Jacobsen, J.V. and Gubler, F. (2009) Anatomical and transcriptomic studies of the coleorhiza reveal the importance of this tissue in regulating dormancy in barley. Plant Physiol. 150, 1006–1021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bartlett, J.G. , Alves, S.C. , Smedley, M. , Snape, J.W. and Harwood, W.A. (2008) High‐throughput Agrobacterium‐mediated barley transformation. Plant Methods, 4, 22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Benech‐Arnold, R.L. , Gualano, N. , Leymarie, J. , Come, D. and Corbineau, F. (2006) Hypoxia interferes with ABA metabolism and increases ABA sensitivity in embryos of dormant barley grains. J. Exp. Bot. 57, 1423–1430. [DOI] [PubMed] [Google Scholar]
- Bethke, P.C. , Gubler, F. , Jacobsen, J.V. and Jones, R.L. (2004) Dormancy of Arabidopsis seeds and barley grains can be broken by nitric oxide. Planta, 219, 847–855. [DOI] [PubMed] [Google Scholar]
- Bradford, K.J. , Benech‐Arnold, R.L. , Come, D. and Corbineau, F. (2008) Quantifying the sensitivity of barley seed germination to oxygen, abscisic acid, and gibberellin using a population‐based threshold model. J. Exp. Bot. 59, 335–347. [DOI] [PubMed] [Google Scholar]
- Côme, D. and Tissaoui, T. (1968) Induction d'une dormance embryonnaire secondaire chez le pommier (Pirus Malus L.) par des atmosphères très appauvries en oxygène. Comptes Rendus de l'Académie des Sciences, Paris, série D, 266, 477–479. [Google Scholar]
- Everitt, B. (2005) An R and S‐plus Companion to Multivariate Analysis. London: Springer; London. [Google Scholar]
- Frottin, F. , Martinez, A. , Peynot, P. , Mitra, S. , Holz, R.C. , Giglione, C. and Meinnel, T. (2006) The proteomics of N‐terminal methionine cleavage. Mol. Cell Proteomics, 5, 2336–2349. [DOI] [PubMed] [Google Scholar]
- Fukao, T. , Xu, K.N. , Ronald, P.C. and Bailey‐Serres, J. (2006) A variable cluster of ethylene response factor‐like genes regulates metabolic and developmental acclimation responses to submergence in rice(W). Plant Cell, 18, 2021–2034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fukao, T. , Yeung, E. and Bailey‐Serres, J. (2011) The submergence tolerance regulator SUB1A mediates crosstalk between submergence and drought tolerance in rice. Plant Cell, 23, 412–427. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fukao, T. , Yeung, E. and Bailey‐Serres, J. (2012) The submergence tolerance gene SUB1A Delays leaf senescence under prolonged darkness through hormonal regulation in rice. Plant Physiol. 160, 1795–1807. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Garzon, M. , Eifler, K. , Faust, A. , Scheel, H. , Hofmann, K. , Koncz, C. and Bachmair, A. (2007) PRT6/At5 g02310 encodes an Arabidopsis ubiquitin ligase of the N‐end rule pathway with arginine specificity and is not the CER3 locus. FEBS Lett. 581, 3189–3196. [DOI] [PubMed] [Google Scholar]
- Gibbs, D.J. , Lee, S.C. , Isa, N.M. , Gramuglia, S. , Fukao, T. , Bassel, G.W. and Holdsworth, M.J. (2011) Homeostatic response to hypoxia is regulated by the N‐end rule pathway in plants. Nature, 479, 415–418. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gibbs, D.J. , Bacardit, J. , Bachmair, A. and Holdsworth, M.J. (2014a) The eukaryotic N‐end rule pathway: conserved mechanisms and diverse functions. Trends Cell Biol. 24, 603–611. [DOI] [PubMed] [Google Scholar]
- Gibbs, D.J. , Isa, N.M. , Movahedi, M. , Lozano‐Juste, J. , Mendiondo, G.M. , Berckhan, S. and Holdsworth, M.J. (2014b) Nitric Oxide sensing in plants is mediated by proteolytic control of group VII ERF transcription factors. Mol. Cell, 53, 369–379. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goldberg, K.M. and Iglewicz, B. (1992) Bivariate extensions of the boxplot. Technometrics, 34, 307–320. [Google Scholar]
- Graciet, E. , Walter, F. , O'Maoileidigh, D. , Pollmann, S. , Meyerowitz, E.M. , Varshavsky, A. and Wellmer, F. (2009) The N‐end rule pathway controls multiple functions during Arabidopsis shoot and leaf development. Proc. Natl. Acad. Sci. USA, 106, 13618–13623. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Graciet, E. , Mesiti, F. and Wellmer, F. (2010) Structure and evolutionary conservation of the plant N‐end rule pathway. Plant J. 61, 741–751. [DOI] [PubMed] [Google Scholar]
- Gubler, F. , Hughes, T. , Waterhouse, P. and Jacobsen, J. (2008) Regulation of dormancy in barley by blue light and after‐ripening: effects on abscisic acid and gibberellin metabolism. Plant Physiol. 147, 886–896. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hattori, Y. , Nagai, K. , Furukawa, S. , Song, X.J. , Kawano, R. , Sakakibara, H. and Ashikari, M. (2009) The ethylene response factors SNORKEL1 and SNORKEL2 allow rice to adapt to deep water. Nature, 460, 1026–U1116. [DOI] [PubMed] [Google Scholar]
- Hoang, H.H. , Bailly, C. , Corbineau, F. and Leymarie, J. (2013) Induction of secondary dormancy by hypoxia in barley grains and its hormonal regulation. J. Exp. Bot. 64, 2017–2025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Holman, T.J. , Jones, P.D. , Russell, L. , Medhurst, A. , Tomas, S.U. , Talloji, P. and Holdsworth, M.J. (2009) The N‐end rule pathway promotes seed germination and establishment through removal of ABA sensitivity in Arabidopsis. Proc. Natl. Acad. Sci. USA, 106, 4549–4554. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hu, R.G. , Sheng, J. , Qi, X. , Xu, Z.M. , Takahashi, T.T. and Varshavsky, A. (2005) The N‐end rule pathway as a nitric oxide sensor controlling the levels of multiple regulators. Nature, 437, 981–986. [DOI] [PubMed] [Google Scholar]
- Jarosova, J. and Kundu, J.K. (2010) Validation of reference genes as internal control for studying viral infections in cereals by quantitative real‐time RT‐PCR. BMC Plant Biol. 10, doi:10.1186/1471‐2229‐10‐146. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jung, J. , Won, S.Y. , Suh, S.C. , Kim, H. , Wing, R. , Jeong, Y. and Kim, M. (2007) The barley ERF‐type transcription factor HvRAF confers enhanced pathogen resistance and salt tolerance in Arabidopsis. Planta, 225, 575–588. [DOI] [PubMed] [Google Scholar]
- Kurowska, M. , Daszkowska‐Golec, A. , Gruszka, D. , Marzec, M. , Szurman, M. , Szarejko, I. and Maluszynski, M. (2011) TILLING ‐ a shortcut in functional genomics. J. Appl. Genet. 52, 371–390. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee, M.J. , Tasaki, T. , Moroi, K. , An, J.Y. , Kimura, S. , Davydov, I.V. and Kwon, Y.T. (2005) RGS4 and RGS5 are in vivo substrates of the N‐end rule pathway. Proc. Natl. Acad. Sci. USA, 102, 15030–15035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li, H.B. , Vaillancourt, R. , Mendham, N. and Zhou, M.X. (2008) Comparative mapping of quantitative trait loci associated with waterlogging tolerance in barley (Hordeum vulgare L.). BMC Genom. 2009, 1:115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li, W.T. , Liu, C.J. , Liu, Y.X. , Pu, Z.E. , Dai, S.F. , Wang, J.R. and Wei, Y.M. (2013) Meta‐analysis of QTL associated with tolerance to abiotic stresses in barley. Euphytica, 189, 31–49. [Google Scholar]
- Licausi, F. , van Dongen, J.T. , Giuntoli, B. , Novi, G. , Santaniello, A. , Geigenberger, P. and Perata, P. (2010) HRE1 and HRE2, two hypoxia‐inducible ethylene response factors, affect anaerobic responses in Arabidopsis thaliana. Plant J. 62, 302–315. [DOI] [PubMed] [Google Scholar]
- Licausi, F. , Kosmacz, M. , Weits, D.A. , Giuntoli, B. , Giorgi, F.M. , Voesenek, L.A.C.J. and van Dongen, J.T. (2011) Oxygen sensing in plants is mediated by an N‐end rule pathway for protein destabilization. Nature, 479, 419–422. [DOI] [PubMed] [Google Scholar]
- Mano, Y. and Takeda, K. (2012) Accurate evaluation and verification of varietal ranking for flooding tolerance at the seedling stage in barley (Hordeum vulgare L.). Breed. Sci. 62, 3–10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mayer, K.F.X. , Waugh, R. , Langridge, P. , Close, T.J. , Wise, R.P. , Graner, A. and Int Barley Genome Sequencing, C . (2012) A physical, genetic and functional sequence assembly of the barley genome. Nature, 491, 711–716. [DOI] [PubMed] [Google Scholar]
- Mendiondo, G.M. , Leymarie, J. , Farrant, J.M. , Corbineau, F. and Benech‐Arnold, R.L. (2010) Differential expression of abscisic acid metabolism and signalling genes induced by seed‐covering structures or hypoxia in barley (Hordeum vulgare L.) grains. Seed Sci. Res. 20, 69–77. [Google Scholar]
- Mendiondo, G.M. , Medhurst, A. , van Roermund, C.W. , Zhang, X. , Devonshire, J. , Scholefield, D. and Holdsworth, M.J. (2014) Barley has two peroxisomal ABC transporters with multiple functions in β‐oxidation. J. Exp. Bot. 65, 4833–4847. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mustroph, A. , Zanetti, M.E. , Jang, C.J.H. , Holtan, H.E. , Repetti, P.P. , Galbraith, D.W. and Bailey‐Serres, J. (2009) Profiling translatomes of discrete cell populations resolves altered cellular priorities during hypoxia in Arabidopsis. Proc. Natl. Acad. Sci. USA, 106, 18843–18848. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nakano, T. , Suzuki, K. , Fujimura, T. and Shinshi, H. (2006) Genome‐wide analysis of the ERF gene family in Arabidopsis and rice. Plant Physiol. 140, 411–432. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nicot, N. , Hausman, J.F. , Hoffmann, L. and Evers, D. (2005) Housekeeping gene selection for real‐time RT‐PCR normalization in potato during biotic and abiotic stress. J. Exp. Bot. 56, 2907–2914. [DOI] [PubMed] [Google Scholar]
- Ogawa, M. , Hanada, A. , Yamauchi, Y. , Kuwalhara, A. , Kamiya, Y. and Yamaguchi, S. (2003) Gibberellin biosynthesis and response during Arabidopsis seed germination. Plant Cell, 15, 1591–1604. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Osnato, M. , Stile, M.R. , Wang, Y.M. , Meynard, D. , Curiale, S. , Guiderdoni, E. and Rossini, L. (2010) Cross Talk between the KNOX and ethylene pathways is mediated by intron‐binding transcription factors in barley. Plant Physiol. 154, 1616–1632. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pang, J.Y. , Zhou, M.X. , Mendham, N. and Shabala, S. (2004) Growth and physiological responses of six barley genotypes to waterlogging and subsequent recovery. Aust. J. Agric. Res. 55, 895–906. [Google Scholar]
- Porra, R.J. , Thompson, W.A. and Kriedemann, P.E. (1989) Determination of accurate extinction coefficients and simultaneous‐equations for assaying chlorophyll‐a and chlorophyll‐b extracted with 4 different solvents ‐ verification of the concentration of chlorophyll standards by atomic‐absorption spectroscopy. Biochim. Biophys. Acta, 975, 384–394. [Google Scholar]
- RDevelopmentCoreTeam (2012) R: A Language and Environment for Statistical Computing. Vienna, Austria: R Foundation for Statistical Computing. [Google Scholar]
- Ross, S. , Giglione, C. , Pierre, M. , Espagne, C. and Meinnel, T. (2005) Functional and developmental impact of cytosolic protein N‐terminal methionine excision in arabidopsis. Plant Physiol. 137, 623–637. [DOI] [PMC free article] [PubMed] [Google Scholar]
- RStudio (2012) RStudio: Integrated Development Environment for R (Version 0.95.262). Boston, MA: R Studio. [Google Scholar]
- de San Celedonio, R.P. , Abeledo, L.G. and Miralles, D.J. (2014) Identifying the critical period for waterlogging on yield and its components in wheat and barley. Plant Soil, 378, 265–277. [Google Scholar]
- Schindelin, J. , Arganda‐Carreras, I. , Frise, E. , Kaynig, V. , Longair, M. , Pietzsch, T. and Cardona, A. (2012) Fiji: an open‐source platform for biological‐image analysis. Nat. Methods, 9, 676–682. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Setter, T.L. and Waters, I. (2003) Review of prospects for germplasm improvement for waterlogging tolerance in wheat, barley and oats. Plant Soil, 253, 1–34. [Google Scholar]
- Till, B.J. , Zerr, T. , Comai, L. and Henikoff, S. (2006) A protocol for TILLING and Ecotilling in plants and animals. Nat. Protoc. 1, 2465–2477. [DOI] [PubMed] [Google Scholar]
- Varshavsky, A. (2011) The N‐end rule pathway and regulation by proteolysis. Protein Sci. 20, 1298–1345. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vashisht, D. , Hesselink, A. , Pierik, R. , Ammerlaan, J.M.H. , Bailey‐Serres, J. , Visser, E.J.W. and Sasidharan, R. (2011) Natural variation of submergence tolerance among Arabidopsis thaliana accessions. New Phytol. 190, 299–310. [DOI] [PubMed] [Google Scholar]
- van Veen, H. , Mustroph, A. , Barding, G.A. , Vergeer‐van Eijk, M. , Welschen‐Evertman, R.A.M. , Pedersen, O. and Sasidharan, R. (2013) Two rumex species from contrasting hydrological niches regulate flooding tolerance through distinct mechanisms. Plant Cell, 25, 4691–4707. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Voesenek, L.A.C.J. and Bailey‐Serres, J. (2013) Flooding tolerance: O‐2 sensing and survival strategies. Curr. Opin. Plant Biol. 16, 647–653. [DOI] [PubMed] [Google Scholar]
- Vogel, J.P. , Garvin, D.F. , Mockler, T.C. , Schmutz, J. , Rokhsar, D. , Bevan, M.W. and Int Brachypodium, I. (2010) Genome sequencing and analysis of the model grass Brachypodium distachyon. Nature, 463, 763–768. [DOI] [PubMed] [Google Scholar]
- Weil, C.F. (2009) TILLING in grass species. Plant Physiol. 149, 158–164. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weits, D.A. , Giuntoli, B. , Kosmacz, M. , Parlanti, S. , Hubberten, H.M. , Riegler, H. and Licausi, F. (2014) Plant cysteine oxidases control the oxygen‐dependent branch of the N‐end‐rule pathway. Nat. Commun. doi:10.1038/ncomms4425 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu, K. , Xu, X. , Fukao, T. , Canlas, P. , Maghirang‐Rodriguez, R. , Heuer, S. and Mackill, D.J. (2006) Sub1A is an ethylene‐response‐factor‐like gene that confers submergence tolerance to rice. Nature, 442, 705–708. [DOI] [PubMed] [Google Scholar]
- Xu, Z.S. , Xia, L.Q. , Chen, M. , Cheng, X.G. , Zhang, R.Y. , Li, L.C. and Ma, Y.Z. (2007) Isolation and molecular characterization of the Triticum aestivum L. ethylene‐responsive factor 1 (TaERF1) that increases multiple stress tolerance. Plant Mol. Biol. 65, 719–732. [DOI] [PubMed] [Google Scholar]
- Yamauchi, Y. , Ogawa, M. , Kuwahara, A. , Hanada, A. , Kamiya, Y. and Yamaguchi, S. (2004) Activation of Gibberellin biosynthesis and response pathways by low temperature during imbibition of Arabidopsis thaliana seeds. Plant Cell, 16, 367–378. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu, J. , Hu, S.N. , Wang, J. , Wong, G.K.S. , Li, S.G. , Liu, B. and Yang, H.M. (2002) A draft sequence of the rice genome (Oryza sativa L. ssp indica). Science, 296, 79–92. [DOI] [PubMed] [Google Scholar]
- Zadoks, J.C. , Chang, T.T. and Konzak, C.F. (1974) Decimal code for growth stages of cereals. Weed Res. 14, 415–421. [Google Scholar]
- Zhou, M. (2011) Accurate phenotyping reveals better QTL for waterlogging tolerance in barley. Plant Breed. 130, 203–208. [Google Scholar]
- Zhou, M.X. , Johnson, P. , Zhou, G.F. , Li, C.D. and Lance, R. (2012) Quantitative trait loci for waterlogging tolerance in a barley cross of franklin x YuYaoXiangTian Erleng and the relationship between waterlogging and salinity tolerance. Crop Sci. 52, 2082–2088. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Figure S1 A. Alignment of the predicted protein sequences of Group VII ERFs from Arabidopsis thaliana and Hordeum vulgare and wheat TaERF1.
Figure S2 Alignment of the predicted Barley PRT6 sequence with sequelogues from Human (Homo sapiens), yeast (Saccharomyces cerevicieae) and Arabidopsis thaliana.
Figure S3 Expression of the barley PRT6 sequelogue during normal growth and development.
Figure S4 Image analysis of a green and a yellow leaf patch (1 cm2) for quantification of greenness with hue in degrees.
Figure S5 Hue histograms of individual pixels of a green and a yellow leaf patch.
Figure S6 HSV colour space for visualisation of hue and value. Circles are filled with colours specified by their position within the plane of hue and value at maximum saturation of one.
Figure S7 qRT‐PCR analysis of the influence of the HvPRT6 mutant alleles prt6k and prt6i on HvPRT6 gene expression relative to the WT (Sebastian).
Table S1 Genome location of barley ERFVIIs (XLSX file).
Table S2 Transgene copy number in RNAi transgenic lines (XLSX file).
Table S3 List of oligonucleotides used in QrtPCR analyses (XLSX file).
Data S1 ImageJ plug‐in software for analysis of leaf colour (IJM file).


