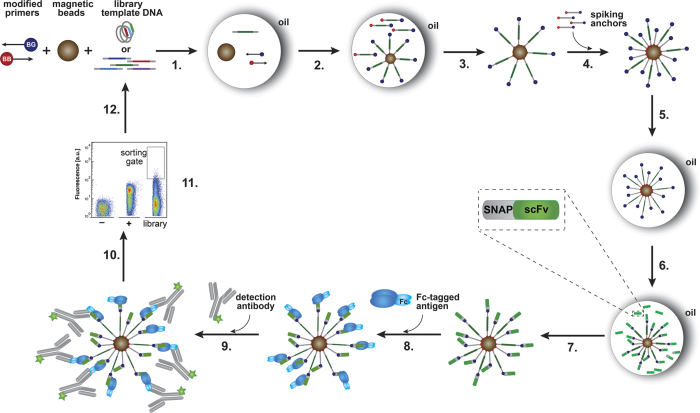
Figure 1. Schematic overview of a selection round using Bead Surface Display (BeSD).
(1) The DNA template, a streptavidin-coated magnetic bead, benzyl guanine (BG)-conjugated primers and biotin (BB)-conjugated primers are encapsulated in water-in-oil droplets, in such a way that the Poisson distribution dictates that there is no more than one DNA template per bead; (2) DNA is amplified by emulsion PCR (ePCR) to give >106 copies, of which ~100–1,000 copies are captured on-bead; (3) the droplet contents are de-emulsified and the beads are washed; (4) BG-BB DNA anchors are added (as additional valencies for display); (5) compartmentalisation of single beads together with IVTT (in vitro transcription/translation) mix in water-in-oil droplets; (6) protein is expressed from the bead-immobilised templates (4 hour expression at 25 °C); (7) de-emulsification liberates beads that are now displaying the protein of interest (e.g., SNAP-scFv-HA), followed by washes to remove the IVTT mixture and unbound excess of expressed protein; (8) incubation with the target (FasR-Fc) followed by washes to remove unbound target; (9) incubation with secondary antibody (anti-Fc DyLight®488); (10) washes to remove excess detection antibody; (11) beads that bind (and show fluorescence above a chosen threshold) are sorted by flow cytometry (FACS) at a rate of ~106 per hour; (12) recovery of the DNA that encoded clones identified as binders. The Figure was adapted from Diamante et al.31 which is available under the terms of the Creative Commons Attribution License (http://creativecommons.org/licenses/by/3.0/).