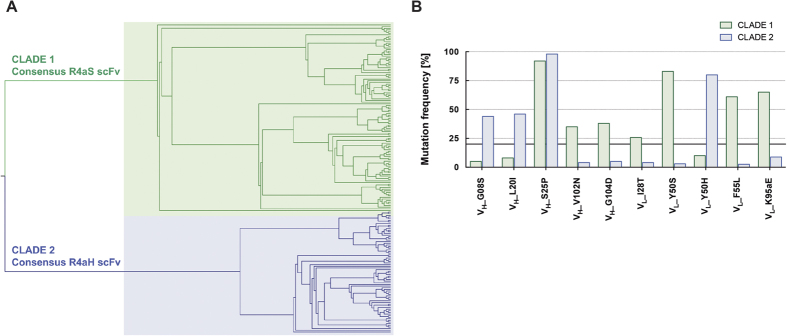
Figure 5. Phylogenetic analysis of the fourth selection round output suggests two consensus designs for R4aS and R4aH.
(A) 193 full-length amino acid sequences from the fourth selection round output scFvs were aligned using the MUSCLE algorithm (online), then the phylogenetic tree was created with CIPRES and visualised in FigTree. The two distinct clades were observed, suggesting that 2 separate lineages of scFvs emerged in the course of the scFv evolution. (B) The graph presents the dominant mutations in the hotspot positions (mutations present in >20% of sequences) in the Clades 1 and 2. The mutations contributing to more than 20% of all analysed sequences in the corresponding clade were classified as consensus mutations. The co-existence of these mutations suggests that no negative epistatic interactions occur between them. Additionally, the fact that they were greatly enriched in the last selection round indicates that they were necessary to improve the target binding properties of the scFvs. The dominant residues for each subsequent clade were assembled into R4aS and R4aH scFv consensus mutants. Interestingly, the scFv containing all consensus mutations from Clade 2 was also identified as the A05a scFv - the most abundant scFv variant in the output of the fourth round of BeSD selection (see Fig. 3C and Table 1), and has ~3-fold improved Kd value (compared to the parent E09 scFv). The second consensus scFv, R4aS, was created (by introducing the relevant mutations to E09 backbone by site-directed mutagenesis), and subsequently its affinity was measured by BLI (see Table 1).