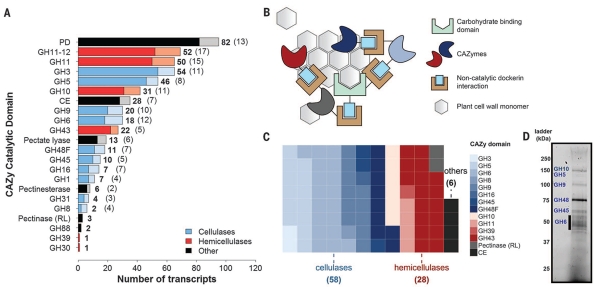
Fig. 2. Anaerobic fungi contain a wealth of biomass-degrading machinery.
(A) Distribution of cellulolytic carbohydrate-active enzyme (CAZy) transcripts and their regulatory antisense transcripts in Piromyces. CAZymes are shown in bold, whereas antisense transcripts are indicated in parentheses and plotted in a lighter shade. In the key, “Other” refers to pectinases and accessory enzymes that separate cellulose and hemicellulose from other cell wall constituents. PD, polysaccharide deacetylase (acetylxylan esterase); CE, carbohydrate esterase (excluding pectinesterases); RL, rhamnogalacturonate lyase. (B) Proposed model for an extracellular catalytic complex for cellulose degradation. (C) CAZyme composition of the putative extracellular complex. Each square represents a single enzyme that encodes a CAZyme fused to at least one dockerin domain. (D) Identity of predominant secreted gut fungal CAZymes in the cellulose-precipitated fraction. Bands were excised and mapped to the transcriptome by tandem MS (fig. S4).