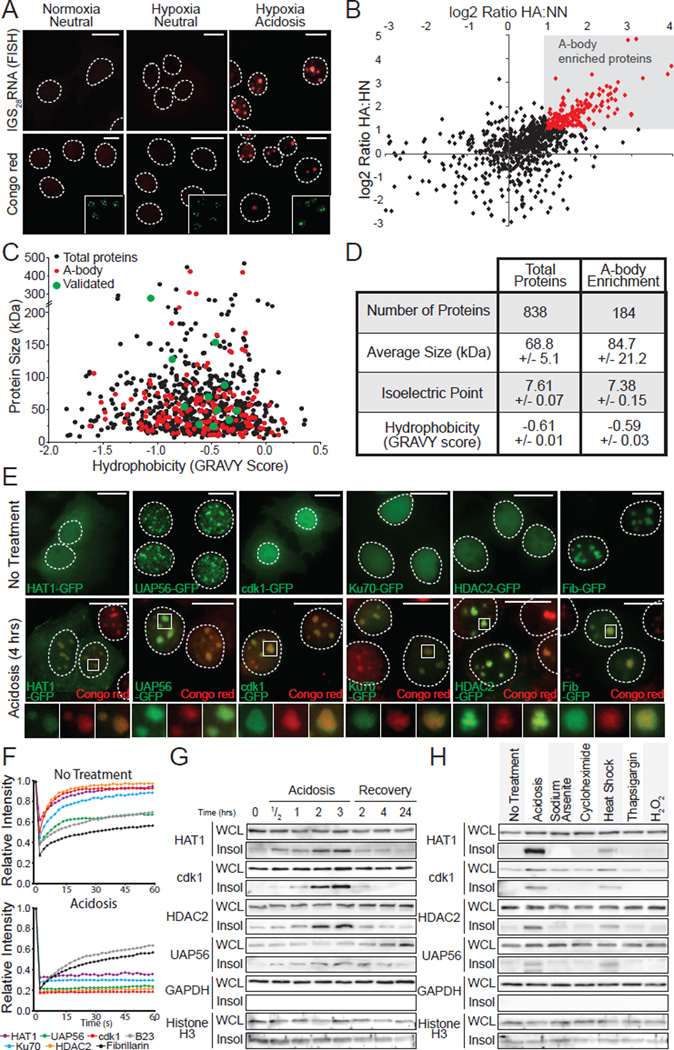
Figure 3. Heterogenic protein composition of the A-body.
(A) Acidosis induces the formation of the A-bodies. MCF-7 cells grown under normoxic-neutral (NN) and hypoxic-neutral (HN) as baseline controls, and hypoxic-acidotic (HA) conditions were stained with Congo red dye or fluorescence in situ hybridized with an anti-sense probe targeting rIGS28RNA. Nucleolar B23 (green) is inset. (B) Identification of the A-body residents. SILAC-MS analysis comparing proteins extracted from MCF-7 cells treated as above. Plot compares the log enrichment of HA:HN versus HA:NN. A-body-enriched (red) proteins fall in the upper right quadrant. (C) A-body components are biochemically similar to the total protein population. Scatter plot of protein size versus hydrophobicity. (D) A heterogeneous family of proteins are targeted to the A-bodies. The size, isoelectric point and hydrophobicity of total and amyloid-specific protein fractions is summarized. Values represent data set averages +/− SEM. (E) Localization of SILAC-MS candidates to the A-bodies. MCF-7 cells transfected with HAT1-GFP, UAP56-GFP, cdk1-GFP, Ku70-GFP, HDAC2-GFP and Fib-GFP were left untreated or exposed to extracellular acidosis prior to staining with Congo red. (F) Targets of the A-bodies are immobilized. Quantification of recovery after photobleaching kinetics for the proteins listed above (E) as the mean relative intensity of at least 5 data sets. (G) SILAC-MS candidates reversibly insolubilize in response to stimuli. MCF-7 cells were exposed to extracellular acidosis and returned to standard growth conditions, for the indicated times prior to harvesting WCL and insoluble proteins. HAT1, cdk1, HDAC2, UAP56, GAPDH and Histone H3 were detected by western blotting. (H) Stimuli-specific insolubilization of SILAC-MS candidates. WCL and insoluble proteins were extracted from untreated, acidotic (2 hrs), heat shocked (2 hrs), sodium arsenite (1 hr), cycloheximide (1hr), thapsigargin (8 hrs) or H2O2 (8 hrs) treated cells. A-body targets/controls were detected as in (E). Dashed circles represent nuclei. Selected regions (white box) were expanded below. White scale bars represents 20mm. See also Figure S3.