ABSTRACT
Bispecific IgG are heterotetramers comprising 2 pairs of heavy and light chains. Co-expression of the 4 component chains in a single host cell typically yields the desired bispecific IgG plus up to 9 additional incorrect chain pairings. Several protein engineering strategies have been reported to facilitate the heterodimerization of antibody heavy chains or cognate pairing of antibody heavy and light chains. These technologies have been used to direct the efficient assembly of bispecific IgG in single host cells and minimize unwanted chain pairings. When purifying bispecific IgGs, the identification and quantification of low levels of closely related IgG contaminants are substantial analytical challenges. Here we have developed a robust high-throughput method for quantitative analysis of bispecific IgG preparations using novel online liquid chromatography in conjunction with an extended mass range Orbitrap-based high-resolution mass spectrometer. A mathematical method was developed to estimate the yields of the 2 isobaric species, namely the desired bispecific IgG and the light chain-scrambled IgG. The analytical methods described herein are anticipated to be broadly applicable to the development of bispecific IgG as drugs and potentially to other complex next-generation biotherapeutics.
KEYWORDS: Bispecific antibody, bispecific IgG quantification, DNA ratios, high-resolution mass spectrometry, single host expression
Abbreviations
- BsIgG
bispecific IgG
- EMR
extended mass range
- ESI
electrospray ionization
- HPLC
high-performance liquid chromatography
- KiH
knobs-into-holes
- IM
ion mobility
- LC-MS
liquid chromatography-mass spectrometry
- MW
molecular weight
- Q-TOF
quadrupole time-of-flight
- RSD
relative standard deviation
- SD
standard deviation
- TIC
total ion chromatogram
Introduction
Bispecific antibodies are of growing interest for drug development, and at least 40 such molecules are currently in clinical studies.1-3 Combining 2 (or more) antigen specificities within a single antibody can endow them with new properties, such as the ability to retarget effector cells to kill tumor cells. Bispecific antibodies can also serve as an alternative, or potentially an improvement, for antibody combination therapies.1,2 Extensive technology development with bispecific antibodies in recent years has led to the generation of at least 60 different alternative formats or scaffolds.1,2,4 The bispecific IgG (BsIgG) format has gained popularity because it may provide IgG-like properties, such as long serum half-life and optional effector functions, as well as the ability to tailor these Fc-associated functions. A BsIgG is a heterotetramer consisting of 2 pairs of heavy and light chains, with each pair providing a different antigen (or epitope) specificity.
Efficient production of BsIgG using a single host cell can be challenging due to promiscuous pairing of the component chains.5 Multiple strategies have been devised to overcome (or avoid) antibody chain pairing problems, as reviewed.2,6 For example, efficient heterodimerization of the 2 heavy chains in BsIgG has been achieved by using the knobs-into-holes (KiH) mutations7,8 and, more recently, by several other elegant strategies.9-12
BsIgG were first produced efficiently in a single host cell using 2 different heavy chains containing KiH mutations in conjunction with a common light chain.13 This strategy circumvents light chain mispairing, but constrains the antibodies that can be used in preparing BsIgG and may require purpose-designed antibody discovery stratagies.14 More recently, separately expressed half-antibodies containing KiH-modified heavy chains and different light chains have been assembled efficiently in vitro.15 More general strategies for assembling BsIgG in single host cells have been developed by engineering antibodies for orthogonal pairing of the 2 light chains to their cognate heavy chains.16-19 For example, a typical design will involve residue modifications at the heavy/light chain interfaces on one or both arms in addition to mutations to facilitate heavy chain heterodimerization.16-18
The success of such antibody engineering designs in facilitating BsIgG assembly can be evaluated following transient coexpression of the component heavy and light chains in mammalian cells. The various IgG species produced are typically purified by protein A or protein G chromatography, and then the BsIgG component of the IgG mixture is quantified by liquid chromatography (LC) in conjunction with mass spectrometry (MS).16-18 Nevertheless, the analytical characterization of BsIgG preparations remains challenging, and new methods are still needed. Native MS and ion mobility (IM) MS are emerging as important tools for the characterization of antibody-based products.20 For example, native MS coupled to size-exclusion chromatography21 and native IM MS22 have been used to analyze BsIgG obtained from the CrossMab technology and antibody-drug conjugates, respectively, under more physiologically representative conditions.
Previously, quadrupole time-of-flight (Q-TOF) LC-MS analyses have been used successfully to measure the relative amounts of different IgG species.23,24 For example, Woods et al.23 coupled a C4 reverse phase LC system with an electrospray ionization (ESI) Q-TOF mass spectrometer to quantify homodimers and associated half-antibody impurities in BsIgG samples. The limit of quantification of antibody impurities was estimated as 2% based upon spiking of standards into purified heterodimer. However, the Q-TOF methodology was not able to resolve IgG species close in mass, impairing sample quantification in some cases. Heck and colleagues have demonstrated quantitative high-resolution analysis of complex mixtures of antibodies by native MS using direct infusion.25-28 The peak width of a single antibody charge state was narrower for an Orbitrap instrument compared to a Q-TOF instrument, which improves the quantification accuracy. Moreover, for the Orbitrap, the centroid of the peak was shifted to slightly lower and closer to the expected mass, due to more efficient desolvation and reduced adduct formation under native conditions.28
Previous work from our lab has demonstrated the benefits of Orbitrap resolution for the identification of unwanted IgG byproducts down to 1% through the use of direct infusion after buffer exchange to either partially denaturing solvents or neutral pH (unpublished data). Although sensitive and effective, this workflow lacked the high-throughput capabilities necessary for large scale evaluation of impurity screening, largely due to the fact that buffer exchange and manual infusion without upfront chromatography can be laborious, thereby limiting the number of samples that can be conveniently analyzed. Furthermore, distinction between the correctly paired BsIgG and the isobaric mispair was not addressed.
Here, we describe an improved platform process for the analysis of BsIgG preparations containing IgG contaminants using reversed phase high-performance (HP) LC coupled with Orbitrap-based high-resolution LC-MS. IgG constructs were engineered to minimize product heterogeneity by deleting the carboxy terminal lysine of the heavy chain (ΔΚ447) and mutation to prevent N-linked Fc glycosylation (N297G). LC conditions and mass spectrometric parameters were optimized to enable the routine analysis of hundreds of BsIgG samples with high reproducibility and sensitivity. A mathematical method was developed to estimate the proportion of BsIgG in an isobaric mixture containing BsIgG and IgG with both light chains mispaired. Lastly, DNA ratios of the component chains used for single cell coexpression were evaluated for their effect on BsIgG production. Taken together, the platform developed here for BsIgG quantification shows exquisite sensitivity and robustness, and has essential utility in evaluating BsIgG designs and in the development of BsIgG therapeutics.
Results
Engineering of the anti-HER2/CD3 BsIgG constructs for minimizing product heterogeneity
An anti-HER2/CD3 BsIgG was constructed from humanized anti-HER2 (humAb4D5-8)29 and anti-CD3 (humAbUCHT1 v9)30 IgG1 antibodies and used as a test system for this study. Knob (T366W) and hole (T366S:L368A:Y407V)7 mutations were installed into the anti-HER2 and anti-CD3 heavy chains, respectively, to promote heavy chain heterodimerization.
IgG samples are typically deglycosylated prior to intact mass analysis by LC-MS. Enzymatic removal of the glycan attached to residue N297 in the CH2 domain of the heavy chain eliminated a major source of mass heterogeneity. This mass heterogeneity was avoided by installing the N297G mutation into the heavy chain of both antibodies, thereby preventing N-linked glycosylation. Another common source of mass heterogeneity in IgG results from proteolysis of the heavy chain C-terminal lysine (K447), from one or both chains, during recombinant IgG production.31 This additional source of mass heterogeneity was circumvented by deleting this lysine residue (ΔK447) from the heavy chain sequences of both the anti-HER2 and anti-CD3 antibodies.
The heavy and light chains for the anti-HER2 and anti-CD3 antibodies were transiently co-transfected at equivalent DNA weight ratios into HEK 293 cells. The secreted IgG was affinity-purified by protein A chromatography from the cell culture conditioned media and analyzed by size-exclusion chromatography. The N297G and ΔK447 heavy chain mutations, alone or in combination, did not affect the transient expression yield of anti-HER2/CD3 BsIgG nor the high proportion of monomeric IgG observed in the size-exclusion chromatography profile (Fig. S1). Therefore, both of these modifications were incorporated into all later constructs so as to minimize the mass heterogeneity for reliable MS analyses.
Detection of IgG using high-resolution LC-MS
In order to quantify IgG species, an HPLC instrument employing a supermacroporous reverse-phase column, MAbPac RP, was coupled to an Exactive Plus extended mass range (EMR) Orbitrap mass spectrometer, and, after optimization, a rapidly obtained, rugged, high quality, baseline-resolved signal was obtained (Fig. 1). The performance of the HPLC system was first evaluated with 30 identical injections of a commercially available IgG mass standard. Reproducibility was significantly improved by replacement of all solvent carrying biocompatible polyetheretherketone lines to stainless steel, and optimization of the composition of the gradient solvents and the column temperature (Fig. S2A, S2B and Table S1). For the same 30 injections, the relative standard deviation (RSD) for major peak parameters (retention time, area, height, width at half height and asymmetry) was greatly reduced (Table S2). The carryover between injections was <0.6% for loading amounts of ≤10 μg IgG (Fig. S2C). Ruggedness, reproducibility and low carry-over were prerequisites for the high throughput screening of large sample sets.
Figure 1.
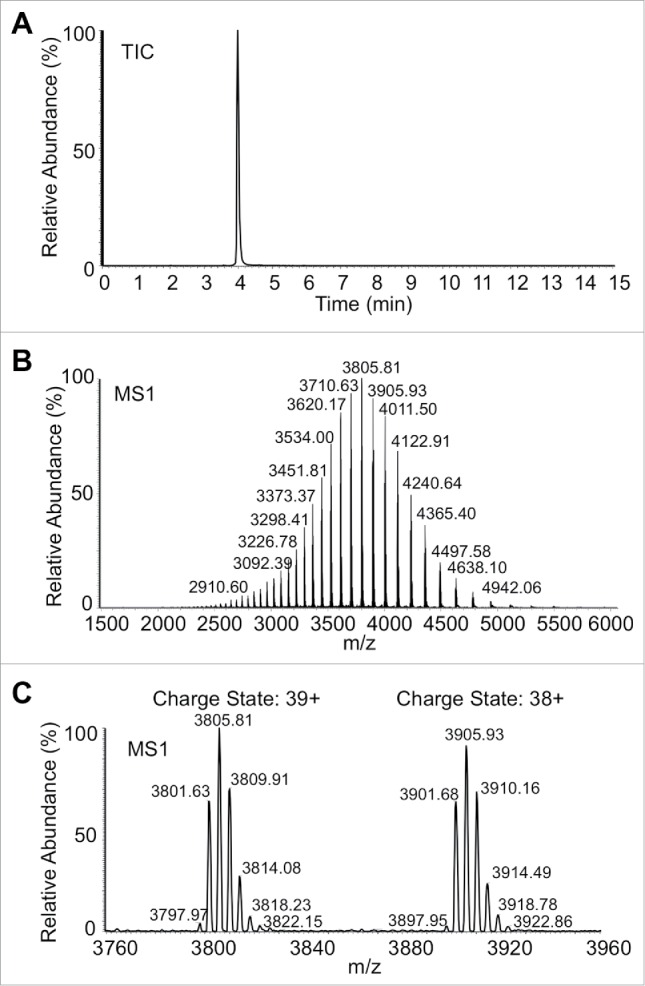
Mass envelope of a glycosylated monoclonal antibody after parameter optimization taken from a 15 min LC-MS analysis under final conditions. (A) TIC trace from 500 ng injected IgG glycosylated standard, (B), ion envelope of (A), and (C) expansion of 2 adjacent charge states of 500 ng IgG standard.
Instrument optimization in Orbitrap Exactive Plus EMR, such as interface condition (sheath gas, Aux gas, S-lens RF level), the desolvation energy (CID, CE) and trapping gas pressure (tapping gas pressure and entrance lens voltage), were performed (see Materials and Methods). A symmetric sharp peak in total ion chromatogram (TIC) with a well-defined ion envelope resulted (Fig. 1A, B). Expansion of 2 adjacent charge states (Fig. 1C) demonstrates baseline resolution of all glycoforms of the glycosylated IgG standard. The mass accuracy with different IgG species was also evaluated, and the average mass error observed was 8 ppm, comparable to previous reports.28
Precise quantification of BsIgG expressed in a single host cell
The quantification ability for this high-resolution LC-MS system was evaluated using BsIgG. For BsIgG production, anti-HER2 and anti-CD3 heavy chains carrying the KiH, N297G, and ΔK447 mutations (H1 and H2) and their respective light chains (L1 and L2) were co-expressed in HEK 293 cells, resulting in 4 major IgG species with minimal contribution from heavy chain homodimerization due to KiH mutations (Fig. 2A). The observed IgG species included the correctly paired anti-HER2/CD3 BsIgG (H1L1/H2L2) and the isobaric light chain-scrambled IgG (H1L2/H2L1). The additional IgG species contained 2 copies of either the anti-HER2 light chain (H1L1/H2L1) or the anti-CD3 light chain (H1L2/H2L2). The mass difference between the BsIgG and either of the IgG containing 2 copies of L1 or L2 reflects the mass difference between the 2 light chains.
Figure 2.
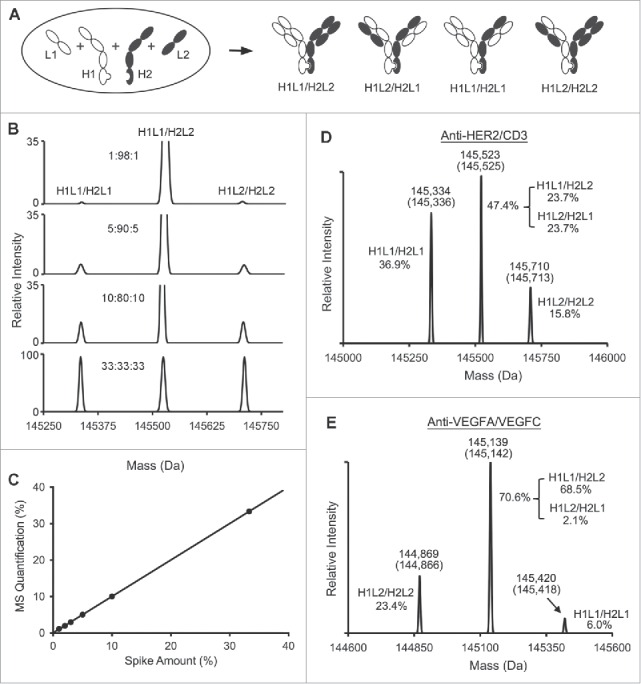
LC-MS quantification of BsIgG samples from expression in single host cells. (A) Schematic diagram of 4 IgG species with heterodimerized heavy chains expressed by co-transfection of HEK 293 cells. (B) Stacked mass spectra of spike-in samples containing anti-HER2/CD3 IgG species, BsIgG (H1L1/H2L2) and 2 mispaired IgG species H1L1/H2L1 and H1L2/H2L2. The heavy and light chains of anti-HER2 are represented as H1 and L1, respectively. The heavy and light chains of anti-CD3 are represented as H2 and L2, respectively. The H1L1/H2L1: H1L1/H2L2: H1L2/H2L2 ratios of 33:33:33, 10:80:10, 5:90:5 and 1:98:1 were analyzed. The BsIgG sample was assembled in vitro using anti-HER2 and anti-CD3 half antibodies,15 and the H1L1/H2L1 and H1L2/H2L2 IgG were produced by co-expression of KiH-modified heavy chains of the anti-HER2 and anti-CD3 antibodies with either of the 2 light chains, L1 or L2, respectively. (C) Plot of measured vs. known percentages of the BsIgG species at different ratios from the spike-in experiment. Data were fitted linearly with an R-squared value of 0.9998. Samples were run in quadruplicate. Refer to table S2 for standard deviations. (D) Mass spectra of the anti-HER2/CD3 BsIgG sample. H1 and L1 represent the heavy and light chains of anti-HER2. H2 and L2 represent the heavy and light chains of anti-CD3. The measured masses and theoretical masses (in parentheses) of each IgG species are labeled on top of each peak. The quantified percentages of the IgG species are also labeled on one side of each peak. (E) Mass spectra of the anti-VEGFA/VEGFC BsIgG sample. H1 and L1 represent the heavy and light chains of anti-VEGFA. H2 and L2 represent the heavy and light chains of anti-VEGFC.
To assess the limits of detection and quantification of the LC-MS platform for BsIgG samples, in vitro assembled anti-HER2/CD3 BsIgG standard (H1L1/H2L2) was titrated with decreasing amounts of 2 mispaired common light chain IgG species (H1L1/H2L1 and H1L2/H2L2) (Fig. 2B and Table S3). For all titrations, the mass spectra showed baseline resolution between the 3 peaks. The close correlation (R2 = 0.9998) between the spiked and measured percentages demonstrated the capability of the optimized LC-MS system to precisely quantify BsIgG species from single-host expressions with contamination levels lower than 1% (Fig. 2C). For all of these data, the standard deviation was below 0.2% (Table S3). Even at the lowest mispair levels, reproducibility was not compromised, which demonstrated the robustness of this platform. As we had demonstrated that the platform enabled the precise quantification of different IgG species in samples with low-level impurities, it was subsequently used to screen panels of BsIgG. Based on the titration experiments, our limits of reproducible quantitation are estimated to be 1%, while the limit of detection (defined as 3 times standard deviation) is 0.3%. Our experience is that peaks below 1% are repeatedly and reliably detected and deconvoluted.
The high-resolution LC-MS system was then employed to analyze the protein A-purified BsIgG samples from single-host expressions. For anti-HER2/CD3 (Fig. 2D and Fig. S3A,C), the peaks of lowest and highest mass represent the common light chain mispaired species, H1L1/H2L1and H1L2/H2L2, respectively. The intermediate peak represents a mixture of the BsIgG and the isobaric light chain-scrambled IgG. The composition of the IgG mixture was measured as 47.4% BsIgG combined with L-chain scrambled IgG, along with 36.9% H1L1/H2L1 and 15.8% H1L2/H2L2.
As an additional example, the expression of an anti-VEGFA/VEGFC BsIgG was assessed (Fig. 2E and Fig. S3B and S3D). Similarly to anti-HER2/CD3, anti-VEGFA and anti-VEGFC heavy chains carrying the KiH, N297G and ΔK447 modifications (H1 and H2) and their respective light chains (L1 and L2) were co-expressed in HEK 293 cells. The desired BsIgG and light chain-scrambled IgG together constituted 70.6%, while the fraction of the IgG with 2 copies of VEGFA or VEGFC light chains were 6.0% and 23.4%, respectively. To accurately assess the efficiency of correct chain pairing, individual percentages of the correctly paired BsIgG and the light chain-scrambled IgG need to be individually determined.
Limits of LC-MS method for resolving different IgG species close in mass
Successful quantification of BsIgG impurities using instrument conditions amenable to high-throughput analysis is dependent on the mass difference between individual antibody arms. To determine the quantification capabilities at a standard operating resolving power (17,500 at 200 m/z), we tested available BsIgG of varying mass differences. Baseline resolution was achieved where antibody impurities differed by 118 Da (Fig. S4A). In contrast, with samples differing in mass by 55 Da, only partial resolution was achieved (Fig. S4B). Specifically, with a 55 Da difference, 2 shoulder peaks were present and amenable to quantification, albeit with reduced accuracy. Due to the fact that high resolving powers require additional spectral acquisition time, they are not compatible with the rapid, efficient chromatographic separation described herein. The peak width of the TIC for a typical LC-MS run was about 0.15 min, which represents 9 data points across the peak (Fig. 1). Increased instrument resolving power (35,000 at m/z 200) on our automated method resulted in 5 useable data points across the peak and overall lower quality results. Additionally, the sensitivity and signal-to-noise ratio was compromised at higher resolving powers, resulting in loss of minor components.
Estimation of the BsIgG and light chain-scrambled IgG content from the MS data
A probability-based mathematical method was developed to estimate the percentage of each of the BsIgG (H1L1/H2L2) and the light chain-scrambled IgG species (H1L2/H2L1) from the combined quantity measured by MS (see Materials and Methods). The anti-HER2/CD3 BsIgG and the corresponding light chain-scrambled species were estimated as 23.7% each (Fig. 2D). For the anti-VEGFA/VEGFC, BsIgG and the light chain scrambled IgG were estimated as 68.5% and 2.1%, respectively (Fig. 2E).
To validate the mathematical model that was used for calculating the component species of the intermediate MS peak, experiments were carried out using the anti-HER2/CD3 and anti-VEGFA/VEGFC BsIgG samples. First, the anti-HER2/CD3 IgG sample was treated with lysyl endopeptidase, which cleaved the heavy chains on the C-terminal side of the K222 residue, located in the upper hinge region. Based on the chain pairing in the IgG sample, 4 Fab species were anticipated in the digested mixture, namely, H1L1, H1L2, H2L1 and H2L2 (Fig. 3A). The H1L1 Fab is derived from BsIgG and the H1L1/H2L1 mispaired IgG. Similarly, the H2L2 Fab is derived from BsIgG and the H1L2/H2L2 IgG. The H1L2 Fab is contributed by both H1L2/H2L2 and H1L2/H2L1 mispair IgG species. The H2L1 Fab was derived from H1L1/H2L1 and H1L2/H2L2 IgG species. The contribution of each Fab species can be calculated from the known IgG content (Fig. 3B, C).
Figure 3.
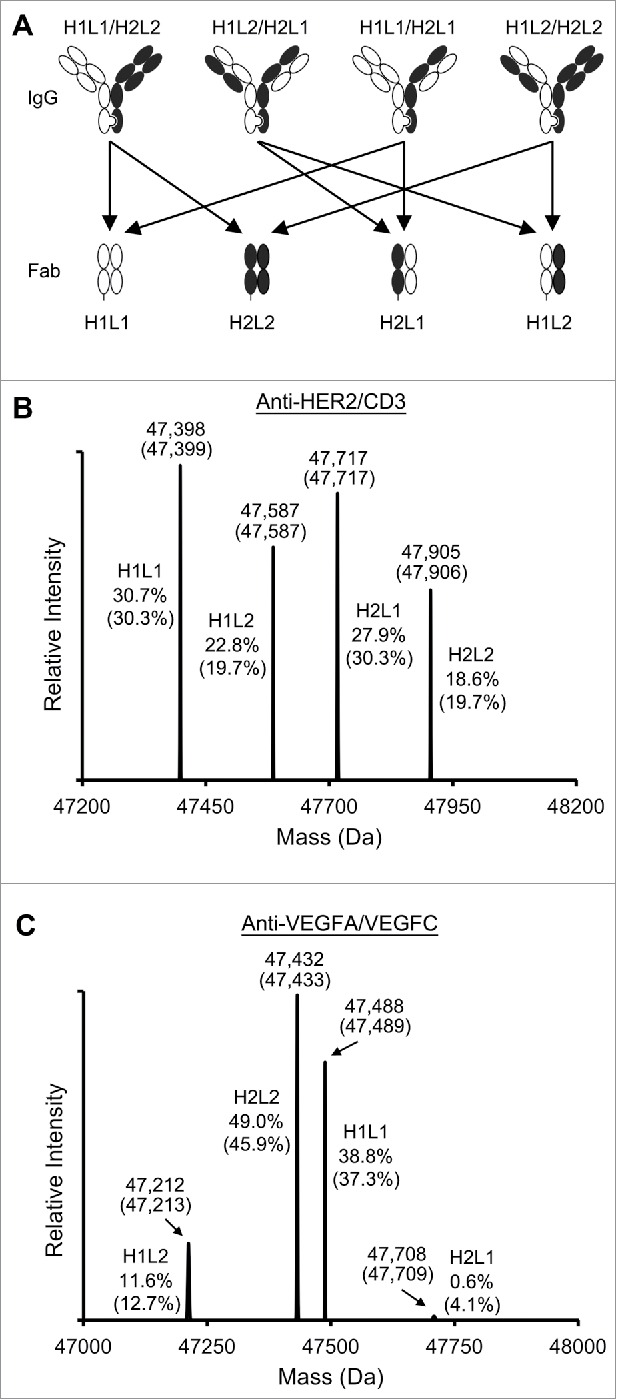
LC-MS analyses of the Fab samples from digested IgG to verify the mathematical estimation of the BsIgG species. (A) Four Fab species derived from lysyl-C digestion of the BsIgG sample. (B) Mass spectra of the digested anti-HER2/CD3 BsIgG sample. H1 and L1 represent the cleaved heavy chain plus light chain of anti-HER2. H2 and L2 represent the cleaved heavy chain plus light chain of anti-CD3. The measured masses and theoretical masses (in parentheses) of each Fab species are labeled on top of each peak. The measured percentages and calculated percentages (in parentheses) of the Fab species are labeled on one side of each peak. (C) Mass spectra of the digested anti-VEGFA/VEGFC BsIgG sample. H1 and L1 represent the cleaved heavy chain and light chain of anti-VEGFA. H2 and L2 represent the cleaved heavy chain and light chain of anti-VEGFC.
To compare with the calculated values, the digested anti-HER2/CD3 BsIgG sample was analyzed using the LC-MS method to quantify the percentages of the Fab fragments (Fig. 3B). In addition, the same experimental method was applied to the anti-VEGFA/VEGFC IgG sample (Fig. 3C). In both cases, the MS-measured percentages closely approximated the calculated compositions of the 4 Fab fragments. Thus, the data supported the use of the mathematical formula for estimating the BsIgG yields from LC-MS measurements.
Effect of DNA chain ratios for co-transfection on the percentage of BsIgG
Multiple weight ratios of anti-HER2/CD3 DNA were tested to study whether the chain ratios have an effect on the percentage of assembled BsIgG. With the total DNA amount for co-transfection being fixed, the L1:L2 ratio was varied while H1:H2 was held constant at 1:1. With the L1:L2 ratios varied from 2.8:1 to 1:2.8, the anti-HER2/CD3 BsIgG content varied over a narrow range: from 19.6% to 24.7% (Fig. 4A, Table S4). The highest BsIgG percentage was observed with an L1:L2 ratio of 1:1.4. In contrast, the percentage of the 2 mispaired common light chain species (H1L1/H2L1 and H1L2/H2L2) varied over more extensive ranges and in a reciprocal manner.
Figure 4.
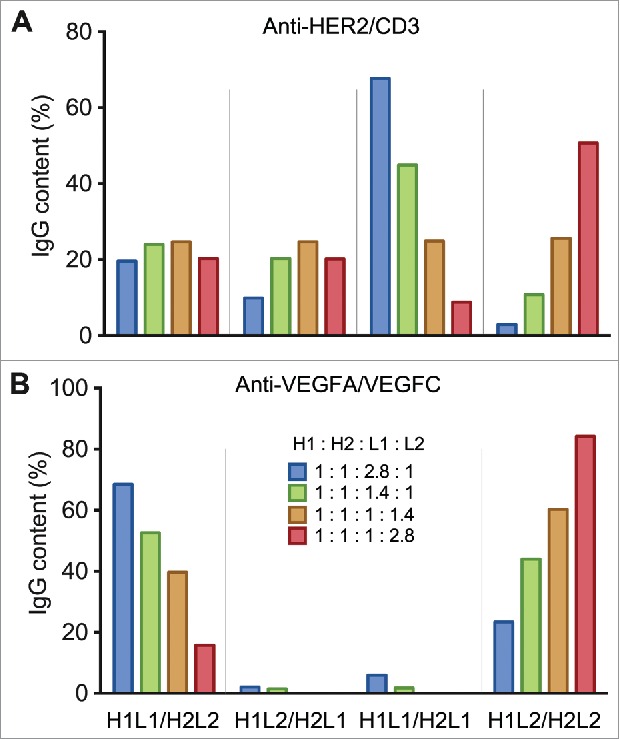
Plots of the percentages of the IgG species (H1L1/H2L2, H1L2/H2L1, H1L1/H2L1, H1L2/H2L2), produced by co-transfection of Expi293 cells with different heavy and light chain DNA weight ratios for (A) anti-HER2/CD3 and (B) anti-VEGFA/VEGFC.
The same DNA chain ratios were also used for production of the anti-VEGFA/VEGFC BsIgG. In this case, the BsIgG yields varied dramatically (from 15.8% to 68.5%) over the different chain ratios (Fig. 4B, Table S5). The percentage of BsIgG was elevated with increasing L1:L2 ratios from 1:2.8 to 2.8:1.
Discussion
As novel therapeutics, bispecific antibodies are of growing interest because they can be used to support and explore new mechanisms of actions for disease treatments. Of all bispecific antibodies, BsIgGs have advantages, and thus are of particular interest, due to their resemblance to conventional IgG therapeutics. However, obtaining pure BsIgG is challenging because of the complexities that arise from the co-expression of 2 pairs of heavy and light chains. Therefore, both the discovery and manufacturing of BsIgG therapeutics benefit from the development of efficient production methods. As the technologies have improved, near-quantitative yields of BsIgG from single host cell expressions have been achieved.7,8,16-18 To confirm the success of engineering solutions, use of a precise and robust companion quantification method for discriminating various engineering designs is critical. For that purpose, we developed a high-resolution LC-MS-based quantification platform compatible with high throughput analytical needs.
Antibodies can be redesigned to improved their homogeneity and their potential for development, as reviewed by Beck et al.32 In this study, humanized anti-HER2 and anti-CD3 antibodies in IgG1 format were modified to avoid 2 sources of heterogeneity. Specifically, the removal of the carbohydrate by the heavy chain N297G mutation eliminated an extra deglycosylation step before LC-MS analyses. Additionally, the deletion of the heavy chain C-terminal lysine residue (ΔK447) circumvented the heterogeneity that may result from proteolytic removal of this lysine.23 As a result, simpler IgG mass spectra could be obtained immediately following protein A chromatography.
Previously, sample analyses for the single-cell expressed BsIgG were mostly performed with the ESI-Q-TOF MS system. By utilizing high-resolution capabilities available on the Orbitrap platform, it was possible to acquire and interpret intra-charge state baseline resolved MS data. With the inclusion of upfront chromatography enabled by a newly designed column demonstrating minimal carry-over (<0.6%) and excellent reproducibility (1.3% RSD for peak area), we quickly, repeatedly and precisely quantified BsIgG impurities under 1% and detected impurities down to 0.3%. This Orbitrap-based high-resolution LC-MS platform performance is superior to the Q-TOF-based LC-MS system due to improved desolvation and increased signal-to-noise ratio. The antibodies used in our study include impurities or modifications that differ by 118 Da or more and are readily and precisely quantified. The higher resolving power necessary to distinguish IgG close in mass, and the concurrent extended spectral acquisition time required, were not compatible with the efficient chromatography methods described herein. In practice, BsIgG combinations that are <100 Da in mass are measured by direct infusion into the mass spectrometer at increased instrument resolution. Barring this complication, our general workflow allows us to determine accurately the distribution of the different IgG species in thousands of BsIgG samples to date at a rate of ∼100 samples per 24-hour period.
Another key aspect of our BsIgG quantification platform is the mathematical method that we developed to estimate the percentages of BsIgG and the light chain-scrambled IgG species. This approach was validated by demonstrating the close approximation between the calculated and experimentally measured values of different Fab fragments obtained by proteolytic digestion of BsIgG mixtures. Two main assumptions were employed in developing these computational methods. First, we assumed that the binding of the light chains to a heavy chain is completely independent from the other heavy chain because it is commonly understood that the 2 Fab arms of an antibody are usually formed independently. Second, we assumed that the BsIgG should have a higher or equal percentage than the light chain-scrambled IgG. This second assumption is consistent with previous reports that some coexpressed antibody pairs show significant preference for pairing of cognate heavy and light chains,16 and other antibody pairs have little (if any) preference for cognate chain pairing.24,33 There are no reports (to our knowledge) of co-expressed antibodies exhibiting a preference for non-cognate heavy and light chain pairing.
In this study, an intrinsic cognate heavy and light chain pairing preference was observed for anti-VEGFA and anti-VEGFC antibodies, resulting in 68.5% yield of BsIgG. In contrast, the heavy and light chains of anti-HER2 and anti-CD3 antibodies paired randomly, giving rise to 24.7% anti-HER2/CD3 BsIgG (Table S4 and S5). Before mathematical correction, these estimates were 70.6% BsIgG for anti-VEGFA/VEGFC and 49.4% BsIgG for anti-HER2/CD3, reflecting a deceptively high estimate of properly paired BsIgG in the latter case. The quantification platform developed here provides a potentially broadly applicable tool to detect and quantify any cognate chain preference when pairs of antibodies are co-expressed.
For the anti-VEGFA/VEGFC and anti-CD3/HER2 BsIgG, the constant domains (CL and CH1) are identical for each antibody pair. Therefore, differences in the variable domain (VL and VH) sequences presumably account for the observed preferential cognate chain pairing for anti-VEGFA/VEGFC and random chain pairing for anti-HER2/CD3. Almost all of the variable domain sequence differences between VEGFA and VEGFC antibodies reside in the complementarity-determining regions, with only a few differences in the framework regions (data not shown). Mutational analysis may help identify the residues responsible for the preferential cognate chain pairing, including the relative contributions of framework region and complementarity-determining region residues.
We have no evidence that antibody light chains are able to swap heavy chain partners post secretion. Once the heavy and light chains are assembled, a disulfide bond is formed between them. This interchain disulfide bond likely serves as a kinetic trap that prevents chain exchange. Even after purposefully reducing the light chain/heavy chain disulfide bond, we were unable to find evidence for light chains swapping heavy chain partners.15 In the case of the anti-HER2/anti-CD3 BsIgG, we constructed the forced chain mispairings (anti-HER2 heavy chain with anti-CD3 light chain or anti-HER2 light chain with anti-CD3 heavy chain) and demonstrated that both cognate heavy and light chains are required for antigen binding for HER2 and also CD3 (J. Zhou, unpublished ELISA data).
We also investigated the effect on the BsIgG yield by the DNA ratios of heavy and light chains used for co-transfection. In most of the previously reported studies involving engineering BsIgG,18-20 the light chain ratio was kept non-optimized as 1:1. However, in this study, the anti-VEGFA/VEGFC BsIgG showed a much lower yield (40-50%) with the equal light chain DNA ratio, compared to the optimized yield (∼69%). Therefore, it may be necessary to evaluate multiple light chain DNA ratios to optimize the percentage of a selected BsIgG. This becomes especially important when comparing different designs or evaluating their performance on various antibodies.
The quantification platform developed here offers high reproducibility and robustness, allowing for the rapid analysis of hundreds of clones in an automated fashion. Applications extend beyond evaluating BsIgG yields of different designs, such as screening clones in the development of stable cell lines. Thus, the platform has the potential to be broadly useful in the development of BsIgG therapeutics. Additionally, our platform may be suitable for other applications in the development of next-generation biotherapeutics, including quantification of bispecific antibody-drug conjugates34 or mixtures of antibodies.20
Materials and methods
Constructs
The sequences of the anti-HER2 (huMAb4D5-8),26 anti-CD3 (huMAbUCHT1v9),17 anti-VEGFA33 and anti-VEGFC29,35-37 antibodies were obtained according to earlier publications. The EU numbering scheme for antibody residues is used throughout this manuscript.38 The heavy chains of anti-HER2 and anti-VEGFA were modified with the “knob” mutation (T366W), and the heavy chains of anti-CD3 and anti-VEGFC with the “hole” mutations (T366S:L368A:Y407V).7 The heavy chains were further modified by site-directed mutagenesis to prevent Fc glycosylation (N297G) and to delete the carboxy terminal lysine (ΔK447). All the antibody constructs were cloned as human IgG1 into the pRK5 mammalian expression vectors.
Antibody expression and purification
The plasmids encoding heavy and light chains for making the BsIgG were mixed according to the weight ratios described in the results section. The DNA mixtures were then co-transfected into Expi293F™ cells (Thermo Fisher Scientific). For sizing analysis as the later application, antibody expressions were performed at the 30 mL scale. The IgG species were purified from the supernatant using the MabSelect Sure protein A agarose beads (GE Healthcare Life Sciences) according to the manufacturer's protocol. For MS as the later application, both antibody expression and purification were performed at the 1 mL scale with high throughput methods that were previously reported.39 Total antibody yields were calculated based upon an extinction coefficient of 1.4 at 280 nm using a Nanodrop instrument (Thermo Fisher Scientific).
Gel filtration analysis
Antibody samples (10 μL) were injected on to a 4.6 mm-diameter TSKgel SuperSW3000 size exclusion column (TOSOH Bioscience) on an Infinity 1260 HPLC instrument (Agilent). The samples were eluted with 200 mM K2PO4, 250 mM KCl, pH 7.0 at a flow rate of 0.35 mL/min.
High-resolution LC-MS
An UltiMate 3000 RSLC (Thermo Fisher Scientific) LC system was configured with HPG-3400RS binary gradient pump with a 400 μL static mixer, WPS-3000TRS thermostatted split loop autosampler, TCC-3000RS thermostatted column compartment, and DAD-3000RS diode array detector with a semi-micro flow cell (2.5 μL, 7 mm). Control of the system was via DCMSlink through Xcalibur software also provided by Thermo Fisher Scientific. A unique reversed phase column was designed specifically for the high throughput characterization of antibodies and antibody fragments by HPLC and LC-MS methods. The MabPac RP column (2.1 mm × 50 mm) consisted of a phenyl hydrophobic supermacroporous 4 μm polymeric resin with 1500 Å pores capable of operation over a wide pH range (pH 0-14) and at temperatures of ≤110°C, offering optimal method development flexibility. The HPLC system was optimized for solvent path, gradient solvent composition and column temperature (Table S1). The final optimized MacPac RP run conditions used a flow rate of 300 μL/min and a column temperature of 80°C. A binary pump was used to deliver solvent A (water containing 0.1% formic acid and 0.02% trifluoroacetic acid) and solvent B (90% acetonitrile containing 9.88% H2O plus 0.1% formic acid and 0.02% trifluoroacetic acid) as a gradient of 20% to 65% solvent B over 4.5 min. The solvent was then step-changed to 90% solvent B and held for 6.4 min to clean the column. Finally, the solvent was step-changed to 20% solvent B and held for 4 min for re-equilibration of the column.
For Intact IgG Mass Check Standard, 500 ng sample was injected via auto-sampler for each run. For BsIgG samples, 3 μg sample was auto-injected from a 96-well plate. The HPLC was coupled to a Thermo Exactive Plus EMR Orbitrap instrument (Thermo Fisher Scientific). The IgG samples were analyzed using the following parameters for data acquisition: 3.90 kV spray voltage; 325°C capillary temperature; 100 S-lens RF level; 15 Sheath gas flow rate and 4 AUX gas flow rate in ESI source; 1500 to 6000 m/z scan range; desolvation, in-source CID 100 eV, CE 0; resolution of 17500 at m/z 200; positive polarity; 10 microscans; 3 × 106 AGC target; fixed AGC mode; 0 averaging; 25 V source DC offset; 8 V injection flatapole DC; 7 V inter flatapole lens; 6 V bent flatapole DC; 0 V transfer multipole DC tune offset; 0 V C-trap entrance lens tune offset; and trapping gas pressure setting of 2. Spectra were visualized using Thermo Xcalibur Qual Browser, then mass spectrum deconvolution was performed with Thermo Protein Deconvolution 4.0 under the following parameters: 10 minimum adjacent charges, 95% confidence noise rejection, 1000∼6000 m/z range, 25 ppm mass tolerance, and 5∼100 charge state range. The relative quantification was based on the intensity reported by Protein Deconvolution 4.0 of each individual peak versus total summed intensities.
Calculation of the percentages of the BsIgG and light chain scrambled IgG
To derive the equation for calculating the percentages of BsIgG, %[H1L1/H2L2], and the light chain-scrambled IgG, %[H1L2/H2L1], we designated the probability of each of the heavy-light chain pairing as p[H1L1], p[H2L2], p[H1L2] and p[H2L1]. Since H1L1 and H1L2 share the same heavy chain H1,
| (1) |
and H2L2 and H2L1 share the same heavy chain H2,
| (2) |
Assuming that the pairing of light chains to the knob heavy chain and the hole heavy chain are completely independent events, the population proportion of BsIgG can be calculated as:
| (3) |
where x = %[H1L1/H2L2]; and the population proportion of the light chain-scrambled IgG can be calculated as:
| (4) |
where y = %[H1L2/H2L1].
From the MS quantification (e.g. Fig. 1D–E), the percentages of the 3 peaks are assigned as a = (%[H1L1/H2L2] + %[H1L2/H2L1]), b = %[H1L1/H2L1] and c = %[H1L2/H2L2], respectively. Therefore,
| (5) |
| (6) |
| (7) |
Assuming that the percentage of BsIgG, %[H1L1/H2L2], is larger than or equal to the percentage of light chain-scrambled IgG %[H1L2/H2L1], from equations (1)-(7), the percentage of BsIgG can be calculated as:
and the percentage of the light chain-scrambled IgG as
In some experiments, e.g., anti-HER2/CD3 at H1:H2:L1:L2 ratios of 1:1:1:1.4 and 1:1:1:2.8 (Table S3), the value of may be a negative number. When this is the case, it is manually forced to be zero; then
Preparation of Fab fragments
One hundred μg protein A-purified IgG were incubated at 37°C with 1 mAU of MS grade lysyl endopeptidase (Wako Laboratory Chemicals) in 100 μL 100 mM Tris-HCl, pH 8.0. The reaction was stopped after 1 h by addition of 5 μL 10% acetic acid. Digested samples were then analyzed by high-resolution LC-MS.
Supplementary Material
Disclosure of potential conflicts of interest
YY, GH, JZ, MD, LM, DE, CS, WS and PJC are current or former employees of Genentech, Inc., which develops and markets drugs for profit. This work was funded by Genentech, Inc.. YY and LM were employees of Genentech while this work was conducted. LG is an employee of Thermo Fisher Scientific, the company who manufactures and markets the mass spectrometry instrumentation used.
Acknowledgments
We thank members of the Research Materials Group in the Early Stage Cell Culture department at Genentech for mammalian antibody expressions and the Antibody Production Group in the Antibody Engineering department for protein purification.
References
- 1.Kontermann RE, Brinkmann U. Bispecific antibodies. Drug Discov Today 2015; 20:838-47; PMID:25728220; http://dx.doi.org/ 10.1016/j.drudis.2015.02.008 [DOI] [PubMed] [Google Scholar]
- 2.Spiess C, Zhai Q, Carter PJ. Alternative molecular formats and therapeutic applications for bispecific antibodies. Mol Immunol 2015; 67:95-106; PMID:25637431; http://dx.doi.org/ 10.1016/j.molimm.2015.01.003 [DOI] [PubMed] [Google Scholar]
- 3.Schuurman J, Parren PW. Editorial overview: Special section: New concepts in antibody therapeutics: What's in store for antibody therapy? Curr Opin Immunol 2016; 40:vii-xiii; PMID:27083411; http://dx.doi.org/ 10.1016/j.coi.2016.04.001 [DOI] [PubMed] [Google Scholar]
- 4.Byrne H, Conroy PJ, Whisstock JC, O'Kennedy RJ. A tale of two specificities: bispecific antibodies for therapeutic and diagnostic applications. Trends Biotechnol 2013; 31:621-32; PMID:24094861; http://dx.doi.org/ 10.1016/j.tibtech.2013.08.007 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Carter P. Bispecific human IgG by design. J Immunol Methods 2001; 248:7-15; PMID:11223065; http://dx.doi.org/ 10.1016/S0022-1759(00)00339-2 [DOI] [PubMed] [Google Scholar]
- 6.Klein C, Sustmann C, Thomas M, Stubenrauch K, Croasdale R, Schanzer J, Brinkmann U, Kettenberger H, Regula JT, Schaefer W. Progress in overcoming the chain association issue in bispecific heterodimeric IgG antibodies. MAbs 2012; 4:653-63; PMID:22925968; http://dx.doi.org/ 10.4161/mabs.21379 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Atwell S, Ridgway JB, Wells JA, Carter P. Stable heterodimers from remodeling the domain interface of a homodimer using a phage display library. J Mol Biol 1997; 270:26-35; PMID:9231898; http://dx.doi.org/ 10.1006/jmbi.1997.1116 [DOI] [PubMed] [Google Scholar]
- 8.Ridgway JB, Presta LG, Carter P. 'Knobs-into-holes' engineering of antibody CH3 domains for heavy chain heterodimerization. Protein Eng 1996; 9:617-21; PMID:8844834; http://dx.doi.org/ 10.1093/protein/9.7.617 [DOI] [PubMed] [Google Scholar]
- 9.Strop P, Ho WH, Boustany LM, Abdiche YN, Lindquist KC, Farias SE, Rickert M, Appah CT, Pascua E, Radcliffe T, et al.. Generating bispecific human IgG1 and IgG2 antibodies from any antibody pair. J Mol Biol 2012; 420:204-19; PMID:22543237; http://dx.doi.org/ 10.1016/j.jmb.2012.04.020 [DOI] [PubMed] [Google Scholar]
- 10.Gunasekaran K, Pentony M, Shen M, Garrett L, Forte C, Woodward A, Ng SB, Born T, Retter M, Manchulenko K, et al.. Enhancing antibody Fc heterodimer formation through electrostatic steering effects: applications to bispecific molecules and monovalent IgG. J Biol Chem 2010; 285:19637-46; PMID:20400508; http://dx.doi.org/ 10.1074/jbc.M110.117382 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Moore GL, Bautista C, Pong E, Nguyen DH, Jacinto J, Eivazi A, Muchhal US, Karki S, Chu SY, Lazar GA. A novel bispecific antibody format enables simultaneous bivalent and monovalent co-engagement of distinct target antigens. MAbs 2011; 3:546-57; PMID:22123055; http://dx.doi.org/ 10.4161/mabs.3.6.18123 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Von Kreudenstein TS, Escobar-Carbrera E, Lario PI, D'Angelo I, Brault K, Kelly J, Durocher Y, Baardsnes J, Woods RJ, Xie MH, et al.. Improving biophysical properties of a bispecific antibody scaffold to aid developability: quality by molecular design. MAbs 2013; 5:646-54; PMID:23924797; http://dx.doi.org/ 10.4161/mabs.25632 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Merchant AM, Zhu Z, Yuan JQ, Goddard A, Adams CW, Presta LG, Carter P. An efficient route to human bispecific IgG. Nat Biotechnol 1998; 16:677-81; PMID:9661204; http://dx.doi.org/ 10.1038/nbt0798-677 [DOI] [PubMed] [Google Scholar]
- 14.Sampei Z, Igawa T, Soeda T, Okuyama-Nishida Y, Moriyama C, Wakabayashi T, Tanaka E, Muto A, Kojima T, Kitazawa T, et al.. Identification and multidimensional optimization of an asymmetric bispecific IgG antibody mimicking the function of factor VIII cofactor activity. PLoS One 2013; 8:e57479; PMID:23468998; http://dx.doi.org/ 10.1371/journal.pone.0057479 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Spiess C, Merchant M, Huang A, Zheng Z, Yang NY, Peng J, Ellerman D, Shatz W, Reilly D, Yansura DG, et al.. Bispecific antibodies with natural architecture produced by co-culture of bacteria expressing two distinct half-antibodies. Nat Biotechnol 2013; 31:753-8; PMID:23831709; http://dx.doi.org/ 10.1038/nbt.2621 [DOI] [PubMed] [Google Scholar]
- 16.Lewis SM, Wu X, Pustilnik A, Sereno A, Huang F, Rick HL, Guntas G, Leaver-Fay A, Smith EM, Ho C, et al.. Generation of bispecific IgG antibodies by structure-based design of an orthogonal Fab interface. Nat Biotechnol 2014; 32:191-8; PMID:24463572; http://dx.doi.org/ 10.1038/nbt.2797 [DOI] [PubMed] [Google Scholar]
- 17.Mazor Y, Oganesyan V, Yang C, Hansen A, Wang J, Liu H, Sachsenmeier K, Carlson M, Gadre DV, Borrok MJ, et al.. Improving target cell specificity using a novel monovalent bispecific IgG design. MAbs 2015; 7:377-89; PMID:25621507; http://dx.doi.org/ 10.1080/19420862.2015.1007816 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Liu Z, Leng EC, Gunasekaran K, Pentony M, Shen M, Howard M, Stoops J, Manchulenko K, Razinkov V, Liu H, et al.. A novel antibody engineering strategy for making monovalent bispecific heterodimeric IgG antibodies by electrostatic steering mechanism. J Biol Chem 2015; 290:7535-62; PMID:25583986; http://dx.doi.org/ 10.1074/jbc.M114.620260 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Schaefer W, Regula JT, Bahner M, Schanzer J, Croasdale R, Durr H, Gassner C, Georges G, Kettenberger H, Imhof-Jung S, et al.. Immunoglobulin domain crossover as a generic approach for the production of bispecific IgG antibodies. Proc Natl Acad Sci U S A 2011; 108:11187-92; PMID:21690412; http://dx.doi.org/ 10.1073/pnas.1019002108 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Terral G, Beck A, Cianférani S. Insights from native mass spectrometry and ion mobility-mass spectrometry for antibody and antibody-based product characterization. J Chromatogr B Analyt Technol Biomed Life Sci 2016; pii: S1570-0232(16)30193-3; PMID:27108304; http://dx.doi.org/ 10.1016/j.jchromb.2016.03.044 [DOI] [PubMed] [Google Scholar]
- 21.Haberger M, Leiss M, Heidenreich AK, Pester O, Hafenmair G, Hook M, Bonnington L, Wegele H, Haindl M, Reusch D, et al.. Rapid characterization of biotherapeutic proteins by size-exclusion chromatography coupled to native mass spectrometry. MAbs 2016; 8:331-9; PMID:26655595; http://dx.doi.org/ 10.1080/19420862.2015.1122150 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Debaene F, Boeuf A, Wagner-Rousset E, Colas O, Ayoub D, Corvaia N, Van Dorsselaer A, Beck A, Cianférani S. Innovative native MS methodologies for antibody drug conjugate characterization: High resolution native MS and IM-MS for average DAR and DAR distribution assessment. Anal Chem 2014; 86:10674-83; PMID:25270580; http://dx.doi.org/ 10.1021/ac502593n [DOI] [PubMed] [Google Scholar]
- 23.Woods RJ, Xie MH, Von Kreudenstein TS, Ng GY, Dixit SB. LC-MS characterization and purity assessment of a prototype bispecific antibody. Mabs 2013; 5:711-22; PMID:23884083; http://dx.doi.org/ 10.4161/mabs.25488 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Schaefer W, Volger HR, Lorenz S, Imhof-Jung S, Regula JT, Klein C, Mølhøj M. Heavy and light chain pairing of bivalent quadroma and knobs-into-holes antibodies analyzed by UHR-ESI-QTOF mass spectrometry. MAbs 2016; 8:49-55; PMID:26496506; http://dx.doi.org/ 10.1080/19420862.2015.1111498 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Rosati S, Rose RJ, Thompson NJ, van Duijn E, Damoc E, Denisov E, Makarov A, Heck AJ. Exploring an orbitrap analyzer for the characterization of intact antibodies by native mass spectrometry. Angew Chem Int Ed Engl 2012; 51:12992-6; PMID:23172610; http://dx.doi.org/ 10.1002/anie.201206745 [DOI] [PubMed] [Google Scholar]
- 26.Rosati S, Thompson NJ, Barendregt A, Hendriks LJ, Bakker AB, de Kruif J, Throsby M, van Duijn E, Heck AJ. Qualitative and semiquantitative analysis of composite mixtures of antibodies by native mass spectrometry. Anal Chem 2012; 84:7227-32; PMID:22882109; http://dx.doi.org/ 10.1021/ac301611d [DOI] [PubMed] [Google Scholar]
- 27.Rosati S, van den Bremer ET, Schuurman J, Parren PW, Kamerling JP, Heck AJ. In-depth qualitative and quantitative analysis of composite glycosylation profiles and other micro-heterogeneity on intact monoclonal antibodies by high-resolution native mass spectrometry using a modified Orbitrap. MAbs 2013; 5:917-24; PMID:23995615; http://dx.doi.org/ 10.4161/mabs.26282 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Thompson NJ, Hendriks LJ, de Kruif J, Throsby M, Heck AJ. Complex mixtures of antibodies generated from a single production qualitatively and quantitatively evaluated by native Orbitrap mass spectrometry. MAbs 2014; 6:197-203; PMID:24351421; http://dx.doi.org/ 10.4161/mabs.27126 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Carter P, Presta L, Gorman CM, Ridgway JB, Henner D, Wong WL, Rowland AM, Kotts C, Carver ME, Shepard HM. Humanization of an anti-p185HER2 antibody for human cancer therapy. Proc Natl Acad Sci U S A 1992; 89:4285-9; PMID:1350088; http://dx.doi.org/ 10.1073/pnas.89.10.4285 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Rodrigues ML, Shalaby MR, Werther W, Presta L, Carter P. Engineering a humanized bispecific F(ab')2 fragment for improved binding to T cells. Int J Cancer Suppl 1992; 7:45-50; PMID:1428403 [PubMed] [Google Scholar]
- 31.Dick LW Jr, Qiu D, Mahon D, Adamo M, Cheng KC. C-terminal lysine variants in fully human monoclonal antibodies: investigation of test methods and possible causes. Biotechnol Bioeng 2008; 100:1132-43; PMID:18553400; http://dx.doi.org/ 10.1002/bit.21855 [DOI] [PubMed] [Google Scholar]
- 32.Beck A, Wurch T, Bailly C, Corvaia N. Strategies and challenges for the next generation of therapeutic antibodies. Nat Rev Immunol 2010; 10:345-52; PMID:20414207; http://dx.doi.org/ 10.1038/nri2747 [DOI] [PubMed] [Google Scholar]
- 33.De Lau WB, Heije K, Neefjes JJ, Oosterwegel M, Rozemuller E, Bast BJ. Absence of preferential homologous H/L chain association in hybrid hybridomas. J Immunol 1991; 146:906-14; PMID:1899099 [PubMed] [Google Scholar]
- 34.Li JY, Perry SR, Muniz-Medina V, Wang X, Wetzel LK, Rebelatto MC, Hinrichs MJ, Bezabeh BZ, Fleming RL, Dimasi N, et al.. A Biparatopic HER2-targeting antibody-drug conjugate induces tumor regression in primary models refractory to or ineligible for HER2-targeted therapy. Cancer Cell 2016; 29:117-29; PMID:26766593; http://dx.doi.org/ 10.1016/j.ccell.2015.12.008 [DOI] [PubMed] [Google Scholar]
- 35.Sun LL, Ellerman D, Mathieu M, Hristopoulos M, Chen X, Li Y, Yan X, Clark R, Reyes A, Stefanich E, et al.. Anti-CD20/CD3 T cell-dependent bispecific antibody for the treatment of B cell malignancies. Sci Transl Med 2015; 7:287ra70; PMID:25972002; http://dx.doi.org/ 10.1126/scitranslmed.aaa4802 [DOI] [PubMed] [Google Scholar]
- 36.Presta LG, Chen H, O'Connor SJ, Chisholm V, Meng YG, Krummen L, Winkler M, Ferrara N. Humanization of an anti-vascular endothelial growth factor monoclonal antibody for the therapy of solid tumors and other disorders. Cancer Res 1997; 57:4593-9; PMID:9377574 [PubMed] [Google Scholar]
- 37.Gogineni A, Caunt M, Crow A, Lee CV, Fuh G, van Bruggen N, Ye W, Weimer RM. Inhibition of VEGF-C modulates distal lymphatic remodeling and secondary metastasis. PLoS One 2013; 8:e68755; PMID:23874750; http://dx.doi.org/ 10.1371/journal.pone.0068755 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Edelman GM, Cunningham BA, Gall WE, Gottlieb PD, Rutishauser U, Waxdal MJ. The covalent structure of an entire gammaG immunoglobulin molecule. Proc Natl Acad Sci U S A 1969; 63:78-85; PMID:5257969; http://dx.doi.org/ 10.1073/pnas.63.1.78 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Bos AB, Luan P, Duque JN, Reilly D, Harms PD, Wong AW. Optimization and automation of an end-to-end high throughput microscale transient protein production process. Biotechnol Bioeng 2015; 112:1832-42; PMID:25851051; http://dx.doi.org/ 10.1002/bit.25601 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


