Figure 4.
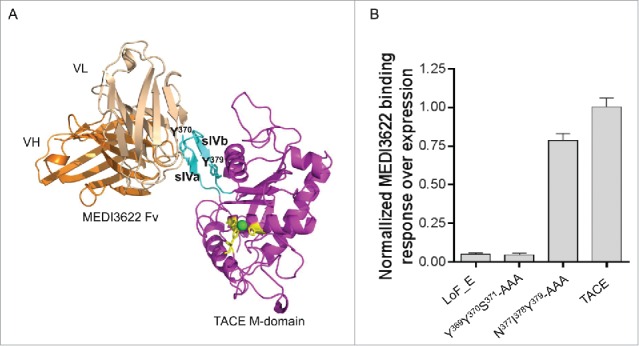
Proposed binding mode of MEDI3622. (A) Three-dimensional model of MEDI3622 bound to TACE. The identified epitope of MEDI3622 (P366-N381) (cyan) adopts a β-hairpin loop conformation projecting from the central body of M-domain (magenta). TACE residues Y370 and Y379 (cyan) along with conserved histidines H405, H409, H415 (yellow) are shown in sticks. The Zn ion is shown as a green sphere. MEDI3622 (VH in orange, VL in beige) binds to TACE through interacting with the sIVa-sIVb β-hairpin loop, with the sIVa strand (Y369Y370S371) located in the center of the binding interface. (B) Binding of MEDI3622 to TACE alanine mutants. Two stretches of amino acids were mutated into alanines, including Y369Y370S371 (sIVa strand, the center of the binding interface) and N377I378Y379 (sIVb strand, peripheral to the binding interface). The alanine variants were expressed as soluble proteins and their binding profiles with MEDI3622 were characterized using SPR. Binding was calculated as % binding compared to wild type TACE after normalization of expression levels using the following formula: [(Response TACE variants MEDI3622/Response TACE wildtype MEDI3622)/(Response TACE variants polyAb/Response TACE wildtype polyAb)] *100. Binding of MEDI3622 to TACE was abolished when mutating residues Y369Y370S371 to alanines, while its binding was retained upon mutating residues N377I378Y379. Results represent the means of 3 independent experiments with error bars indicating standard deviations.
