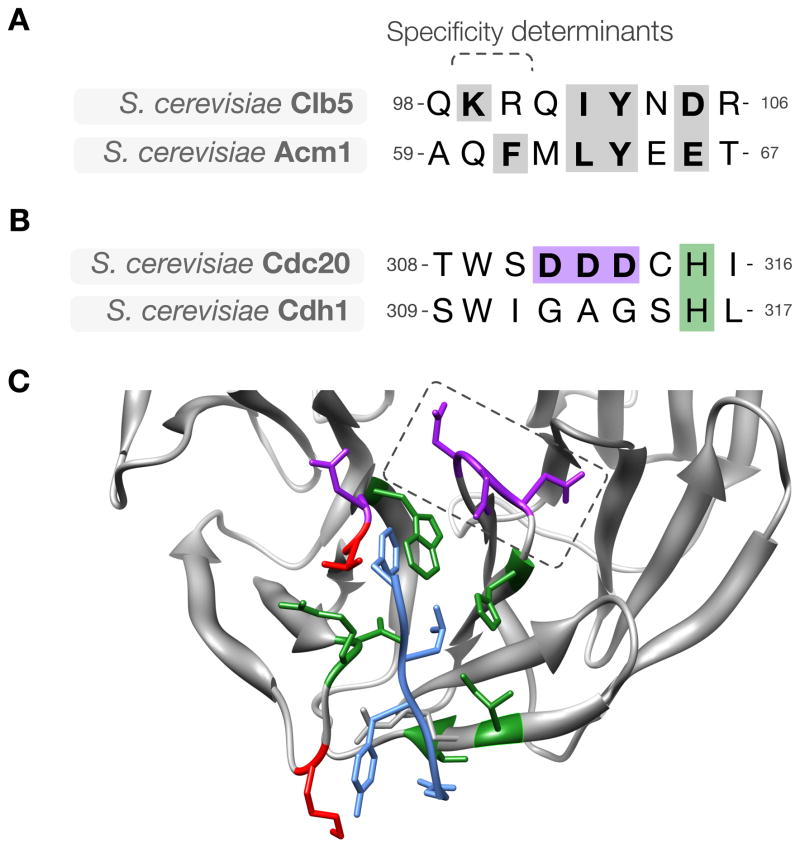
Figure 5. Divergence of ABBA motif binding specificity in evolution.
(A) Alignment of the ABBA motif in budding yeast Clb5 and Acm1, showing key conserved residues (gray). (B) The sequence of the activator region containing the specificity-determining loop in the ABBA motif-binding pocket. The loop is GAG in Cdh1 and DDD in Cdc20. (C) Modeled structure of budding yeast Cdc20 (gray) with the ABBA motif from Acm1 (blue). Pocket residues on Cdc20 are colored based on conservation between budding Cdh1 and Cdc20: green indicates residues conserved between Cdh1 and Cdc20, red indicates residues that are not conserved, and purple indicates Cdc20-specific acidic residues, including the specificity-determining acidic loop (dashed box).