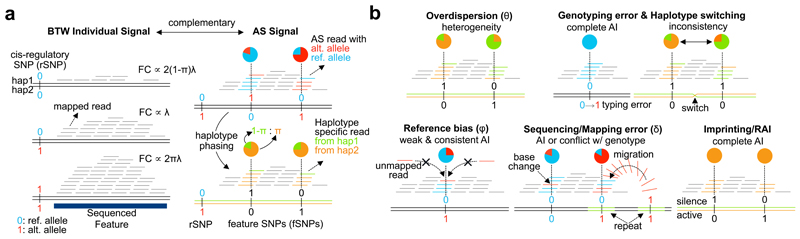
Figure 1.
Schematic of RASQUAL approach. Throughout, reference and alternate alleles are coloured blue and red and coded 0 or 1, respectively, while alternative haplotype are coloured orange and green, respectively. (a) Plot illustrates the two sources of input data to RASQUAL: between-individual and AS signals, as observed from sequence data. Left panel shows the fragment count (FC) is proportional to rSNP genotype and right hand panel illustrates how those two signals are connected by the cis-regulatory effect π after conversion of AS counts into haplotype specific expression (see Main text for details). (b) Visual representation of the key RASQUAL features and parameters. Overdispersion introduces greater heterogeneity in the AS count than would be expected under binomial assumption. RASQUAL models the overdispersion in AS counts and total fragment counts with a single parameter θ. Genotyping error introduces complete allelic imbalance when homozygote is miscalled as heterozygote. Haplotype switching produces inconsistency of allelic imbalance among SNPs within an individual. Reference bias occurs when sequence reads containing the alternative allele(s) are unmappable to the correct location. RASQUAL employs a parameter φ that captures the excess of allelic imbalance beyond the genetic effect π. Sequencing/mapping error introduces additional allelic imbalance or genotype inconsistency. RASQUAL explicitly models the proportion of reads that are erroneously sequenced or mapped from incorrect genomic location by parameter δ to allow imperfect sequencing results. Imprinting introduces strong allelic imbalance that can confounds with genetic effects.