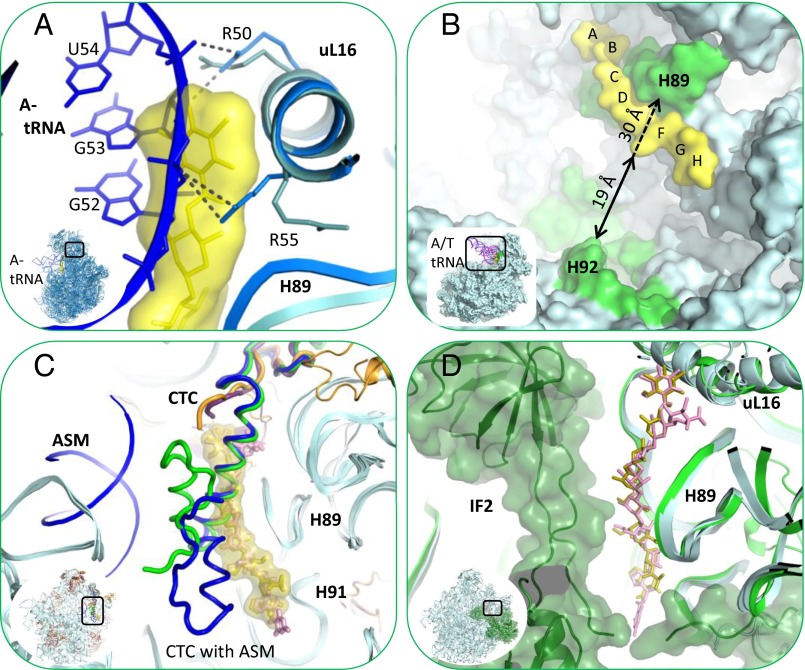
Fig. 5.
Orthosomycins’ inhibition mechanism. Orthosomycin binding pocket overlaps A-tRNA, IF2, and domain 3 of rProtein CTC binding sites and the accommodation corridor. (A) Superposition of A-tRNA (blue) binding site at T70S (marine) crystal structure (PDB ID code 4VD5) with D50S–avi structure (teal, yellow). Avi is shown by surface representation. A-tRNA elbow U54 and G53 nucleotides backbone clash with avi residues A and B. Both avi and the A-tRNA elbow bind to R50 and R55 of rProtein uL16. The side chain of R55 is shifted toward H89 upon avi binding. (B) Accommodation corridor inhibition. A/T tRNA (pink) and A/A tRNA (blue) of E70S EM structure (PDB ID codes 1QZA and 1QZB) superposed on D50S–avi complex structure (teal, yellow). A/T tRNA is accommodated into A site, A/A tRNA, over H92 barrier. Zoom into the accommodation corridor (green), defined by H92 and H89. Avi and evn block the accessibility of accommodation corridor H89 rRNA nucleotides and narrows the accommodation corridor between H89 and H92, from 30 Å (dashed arrow) to 19 Å (black arrow). (C) rProtein CTC (bL25) location and conformation within apo D50S (green) and its complex with ASM (blue) crystal structures (PDB ID codes 1NKW and 1NJM) superposed on D50S–avi crystal structure (teal, yellow). Avi is shown with surface representation. The helix of both CTC conformations clash with avi res B, C, and D, all binding to H89 minor groove. (D) IF2 (dark green) binding site on E70S EM structure (PDB ID codes 3JCJ) superposed on the D50S–avi (teal, yellow) crystal structure. Zoom into IF2 interaction with H89, in proximity to avi and evn binding sites is shown.