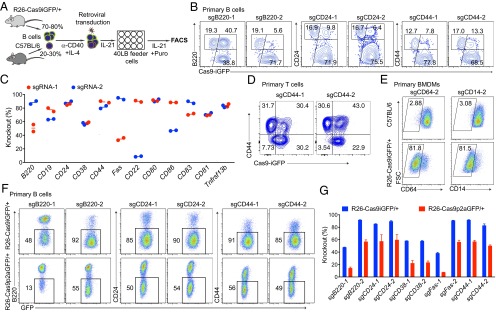
Fig. 2.
Efficient CRISPR-mediated gene inactivation in primary immune cells. (A) Scheme of CRISPR-mediated gene knockout in Cas9-expressing B cells. Splenic B cells isolated from wild-type and Cas9 transgenic mice were mixed at a 1:4 ratio. These cells were activated with anti-CD40 and IL-4 for 2 d and transduced with retroviral particles expressing specific sgRNAs and a puromycin resistance gene. One day after transduction, the cells were transferred into 40LB feeder cell plates and cultured in the presence of IL-21 and puromycin. (B) FACS plots of CD19+BFP+ cells targeted with sgRNAs against B220 (Left), CD24 (Middle), and CD44 (Right) 4 d after transduction. Knockout efficiencies were calculated based on the GFP+ cells that lost the surface marker (subgates of the GFP+ population). (C) Summary of the knockout efficiencies for the 12 indicated surface marker genes with two sgRNAs per gene. Knockout efficiencies were determined by FACS 4 d after transduction of Cas9-expressing primary B cells with the respective sgRNAs. (D) FACS plots showing the CD44 knockout efficiency in Cas9-expressing primary T cells. Mixed Cas9 (GFP+) and wild-type (GFP−) splenic T cells were activated and transduced with retroviral particles expressing sgCD44-1 and sgCD44-2. The gates indicate the knockout efficiency in GFP+ T cells. (E) FACS plots showing the knockout efficiency of CD64 and CD14 in nondividing primary BMDMs. BMDMs from C57BL/6 (Top) and R26-Cas9iGFP/+ (Bottom) mice were transduced with lentiviruses expressing sgCD64-2 and sgCD14-2. (F) FACS plots of knockout efficiencies in primary B cells from R26-Cas9iGFP/+ (Upper) and R26-Cas9p2aGFP/+ (Lower) mice, using the same setup as in A and sgRNAs against B220, CD24, and CD44. (G) Summary of knockout efficiencies using primary B cells from R26-Cas9iGFP/+ (blue) and R26-Cas9p2aGFP/+ (red) mice.