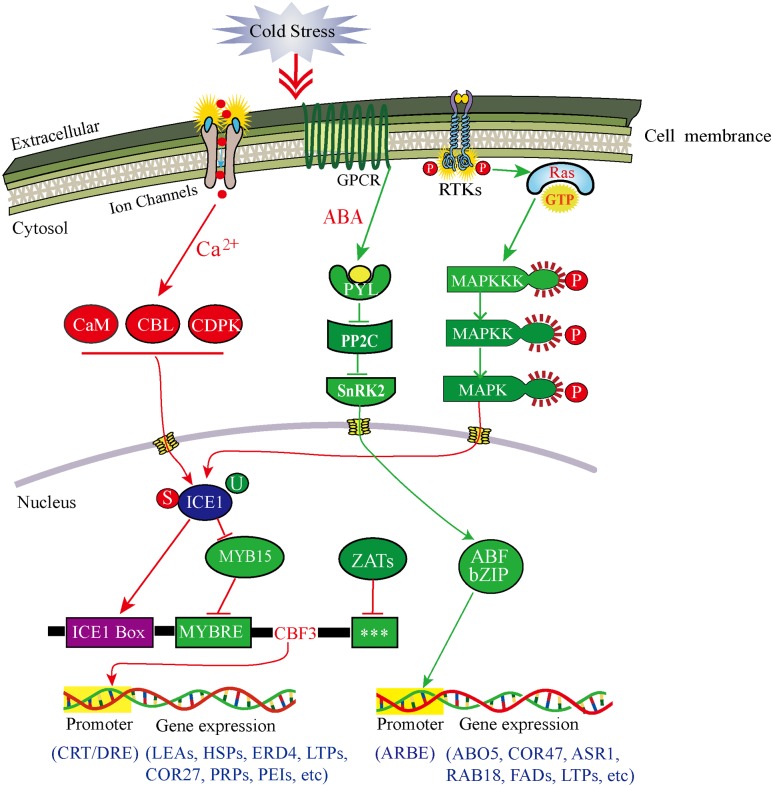
FIGURE 6.
Model of multiple signal transduction pathways and transcriptional networks during cold acclimation in D. officinale. Red lines represent Ca2+-dependent signaling process and CBF-dependent regulatory pathway; green lines represent Ca2+-independent signaling process and ABA-dependent regulator pathway. Arrows represent positive regulation, whereas lines ending with a bar represent negative regulation. The three stars (∗∗∗) indicate unknown cis-elements. ABF, ABA-responsive element binding factor; ABO5, ABA overly- ensitive 5; ABRE, ABF recognition element; ASR1, ABA stress-ripening protein 1; CaM, calmodulin; CBL, calcineurin B-like Ca2+ sensors; CDPK, Ca2+-dependent protein kinase; COR27/47, cold regulated gene 27/47; CRT, C-repeat elements; DRE, dehydration-responsive elements; ERD4, dehydration-induced protein 4; FADs, fatty acid desaturases; GPCR, G-protein-coupled receptor; HSPs, heat shock proteins; LEAs, late embryogenesis abundant protein; LTPs, lipid-transfer proteins; MYBRS, MYB transcription factor recognition sequence; PEIs, pectinesterase inhibitors; PP2C, 2C-type protein phosphatase; PRPs, proline-rich proteins; PYL, pyrabactin resistance 1-like; RAB18, dehydrin family protein 18; RAS, rat sarcoma protein; RTKs, receptor-tyrosine kinases; SnRK2, SNF1-related protein kinase 2; P, phosphorylation; S, sumoylation; U, ubiquitination.