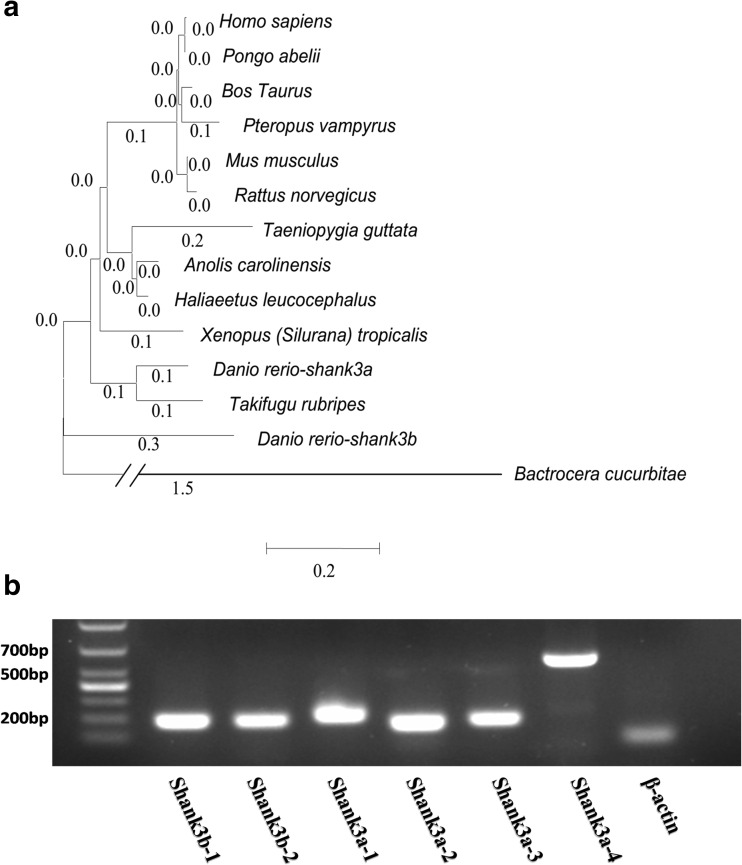
Fig. 2.
Phylogenetic tree of evolutionary relationship of SHANK3 proteins. a Diverse animal phyla were identified by a best reciprocal BLAST search with human SHANK3 and are mapped onto a phylogenetic tree. All of the sequences are available from the NCBI protein database with the exception of zebrafish shank3a, which was obtained from the Ensemble protein database and our study. Homo sapiens (NP_277052.1), Pongo abelii (XP_009232827.1), Rattus norvegicus (NP_067708.1), Mus musculus (NP_067398.2), Bos Taurus (XP_010804153.1), Taeniopygia guttata (XP_012432256.1), Xenopus (Silurana) tropicalis (XP_002941881.2), Danio rerio shank3b (XP_001919745.1), Bactrocera cucurbitae (XP_011184638.1), Pteropus vampyrus (XP_011382480.1), Takifugu rubripes (XP_003967184.1), Anolis carolinensis (XP_008121382.1), Haliaeetus leucocephalus (XP_010564365.1), and Danio rerio shank3a (ENSDART00000139505 and our study). The tree was constructed using the neighbor-joining methods as implemented in the MEGA 6 package. Clade robustness was measured by the bootstrap method with 1000 replicates. The lengths of the lines are proportional to the evolutionary distances from branching points. b Transcript-specific oligonucleotide primers and amplicons were generated to confirm the existence of the key fragment of six predicted transcripts of shank3 in the Ensembl database. cDNA from 3-month-old zebrafish was used as a template for PCR, as noted above each 2 % agarose gel. The housekeeping gene β-actin was used as an internal reference