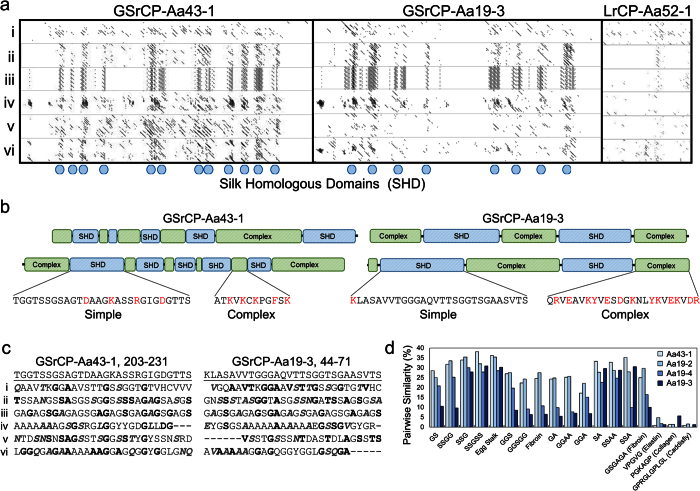
Figure 5. Shared primary structure of polar GSrCPs.
(a) Recursive dot plot analysis of GSrCP-Aa43-1, GSrCP-Aa19-3, and LrCP-Aa52-1 against archetypal silk proteins, highlighting regions of low complexity along primary sequences. (i) AaCP19, (ii) Egg Stalk Silk from M. signata, (iii) Heavy Chain Fibroin from B. mori, (iv) Heavy Chain Fibroin from R. fugax, (v) Sericin I from B. mori, (vi) Spidroin I from N. clavipes. Dots represent silk homologous domains (SHDs). (b) Distinct alternating primary structure observed in GSrCPs defined by alternating silk homologous and complex domains. (c) Pairwise alignment of archetypal silk proteins to representative SHDs, demarked by dot plotting, where bold letters are identical to the query sequence and italicized bold letters are chemically similar. (d) Stringency of primary sequence in GSrCP low complexity regions measured by pairwise alignment to de novo silk-like motifs, showing higher stringency with [SS] based models over [GG]. Percentages are the number of similar and identical alignments between pairs, divided by cement protein sequence length.