Fig. 1. StrigoQuant design, characterization, and analysis of receptor complex requirements.
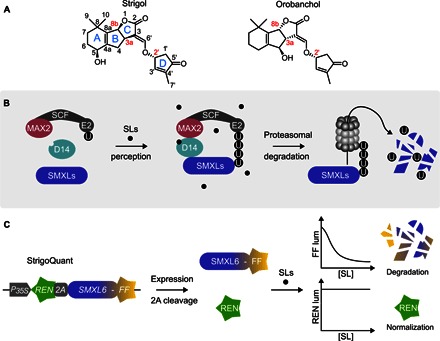
(A) General structure and configuration of the canonical SLs, strigol and orobanchol. Note the stereocenters at the BC-junction (carbons 3a and 8b) and in the D-ring (carbon 2′). (B) Scheme of the SL perception machinery in Arabidopsis. In the presence of SLs, receptor complex formation takes place, with SLs binding to the receptor protein AtD14, with recruitment of SMXLs to the MAX2 and SKP1/CUL1/F-box E2 ubiquitin ligase complex (SCFMAX2). SMXLs are ubiquitinated (U) and consequently degraded by the 26S proteasome. (C) StrigoQuant construct expressing a renilla luciferase (REN; green) connected via a 2A peptide to the sensor module (SM), AtSMXL6 (SMXL6), fused to a firefly luciferase (FF; yellow), under the control of a constitutive 35S promoter. The 2A peptide in the synthetic construct leads to stoichiometric coexpression of REN (normalization element) and SM (SMXL6-FF). Upon the addition of SLs, SMXL6-FF becomes ubiquitinated and degraded, whereas REN expression remains constant, leading to a decrease in the FF/REN ratio.
